-
摘要: 现有的图聚类方法主要存在两方面的问题, 一是对各个类规模一致的假设, 在许多实际应用中并不成立; 二是在处理多类聚类问题时, 其所常借助的递归技术或启发式算法会影响聚类的性能. 为此, 本文提出一种基于灵活平衡约束的多类图聚类方法. 其能够覆盖从绝对平衡约束到无平衡约束的范围, 可同时处理类别规模一致和不一致的问题. 为有效求解新方法中的参数, 进一步提出一个紧松弛方法来使所提出的图聚类方法不仅易于求解, 且在处理多类聚类问题时不必依赖递归技术, 而能直接得到聚类结果. 另外, 本文还给出一种实现松弛图聚类的有效求解算法. 在合成数据和真实数据上的实验结果表明, 所提出的方法具有良好的性能.Abstract: The existing graph clustering methods suffer from two main problems: first, they assume that the size of each class is the same, which is not applicable in many practical applications; second, when dealing with multi-class clustering problems, the recursive technique or heuristic algorithm that is commonly used can affect the clustering performance. To address these issues, this paper proposes a multi-class graph clustering method based on flexible balance constraints. It can cover a range from absolute balance constraints to no balance constraints and can handle both uniform and non-uniform class size problems simultaneously. To effectively solve the parameters in the new method, a tight relaxation method is further proposed to make the proposed graph clustering method not only easy to solve but also able to obtain clustering results directly without relying on recursive techniques when dealing with multi-class clustering problems. In addition, this paper also presents an effective algorithm for implementing the relaxed graph clustering. Experimental results on both synthetic and real data demonstrate that the proposed method has good performance.
-
Key words:
- Graph clustering /
- graph cut /
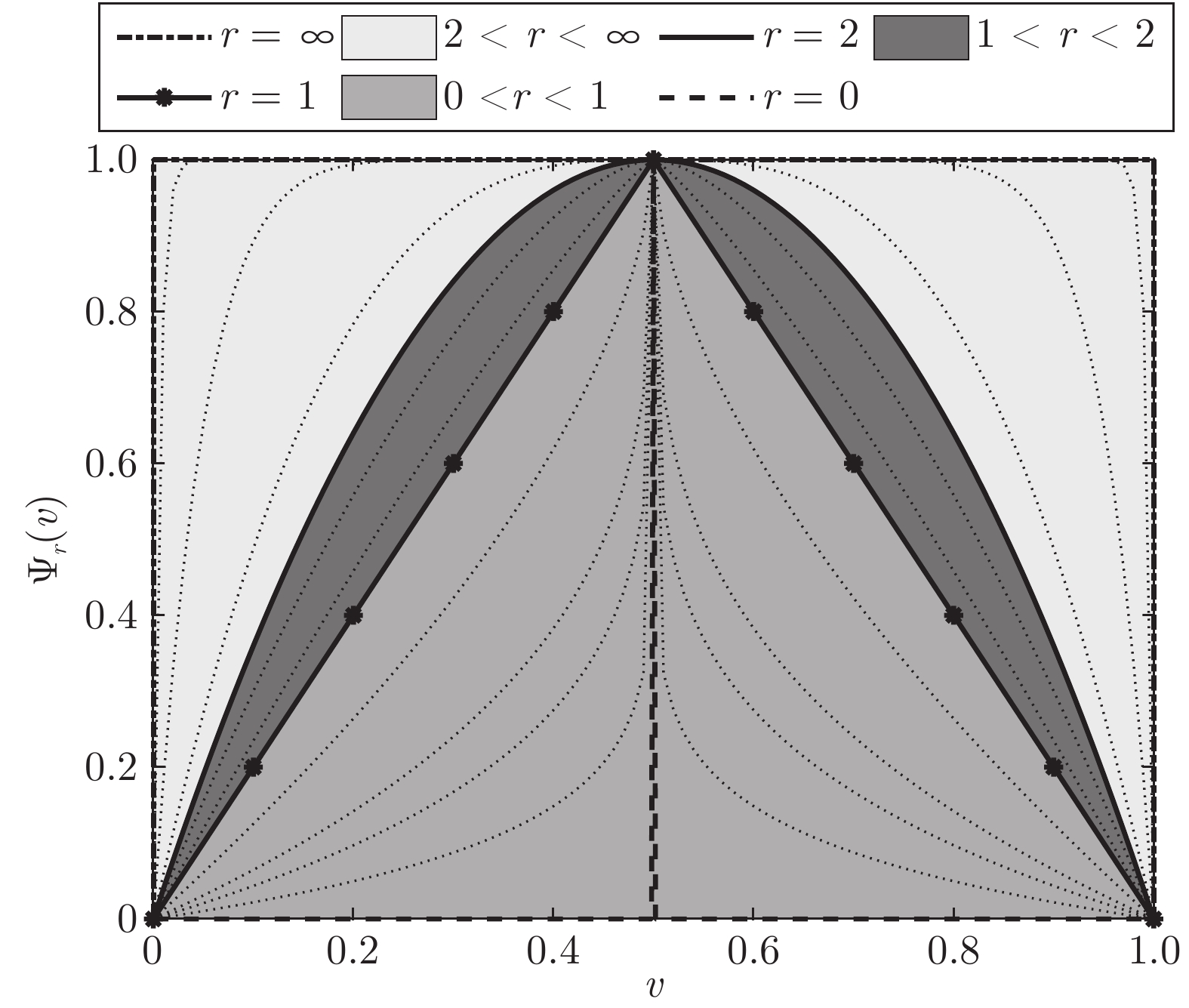
- balanced constraint /
- tight relaxation
-
表 1 真实数据集
Table 1 The statistics of real databases
数据集 样本数 类别数 数据类型 HIGHSCHOOL 60 5 社交 POLBOOKS 105 3 社交 FOOTBALL 115 12 社交 DUKE 44 2 医疗 CANCER 198 14 医疗 KHAN 83 4 基因 ROSETTA 300 5 基因 ORL 400 40 图像 UMIST 575 20 图像 ALPHADIGS 1404 36 图像 COIL20 1440 20 图像 SAVEE 480 7 语音 CASIA 1200 6 语音 IEMOCAP 9994 8 语音 MED 1033 31 文本 WEBKB4 4196 4 文本 REUTERS 8293 65 文本 YEAST 1484 10 生物 FAULTS 1941 7 工业 ECOLI 327 5 蛋白质 表 2 各图聚类方法的聚类精度(%)对比
Table 2 Comparison of clustering accuracies (%) between graph clustering methods
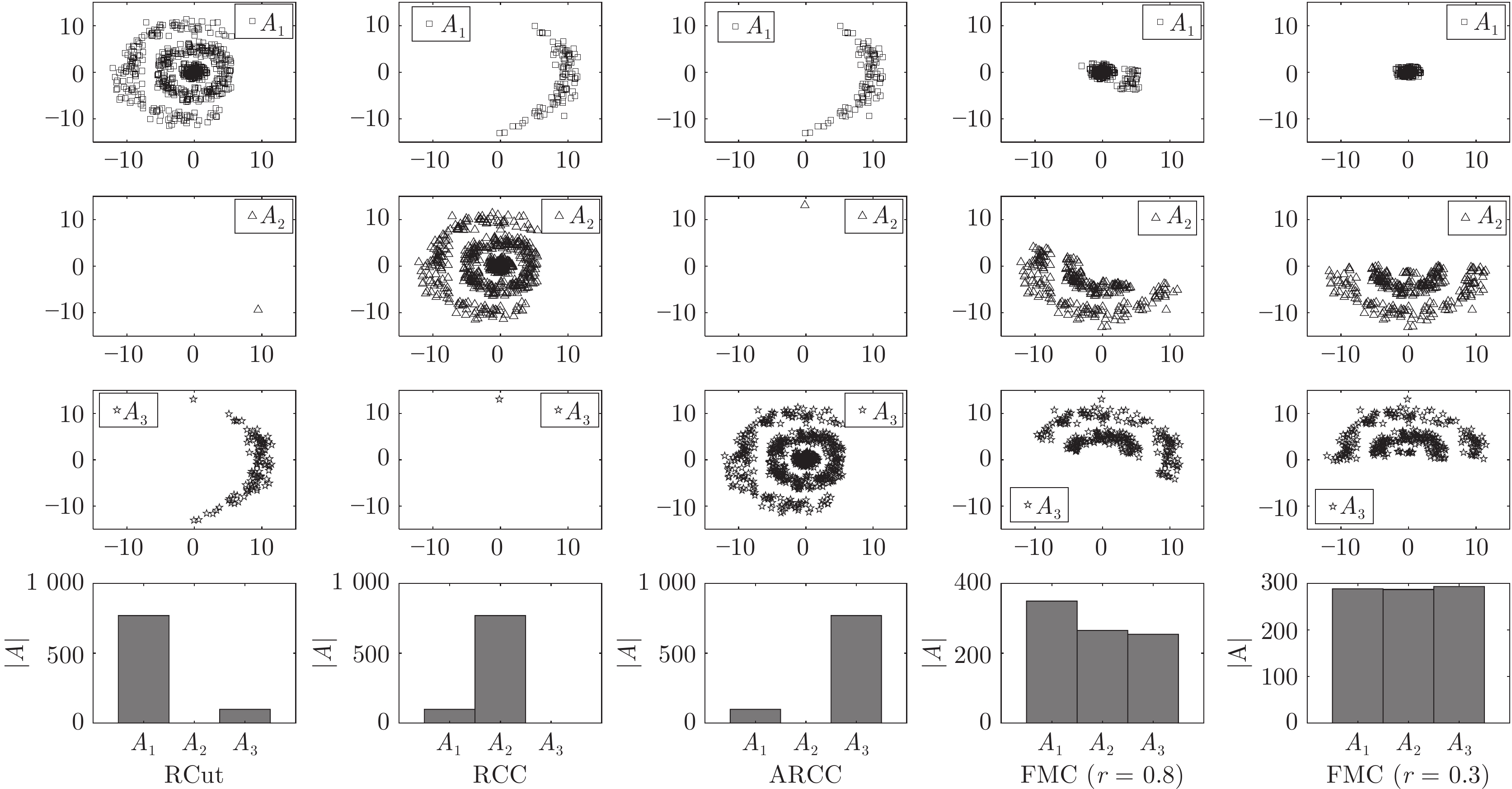
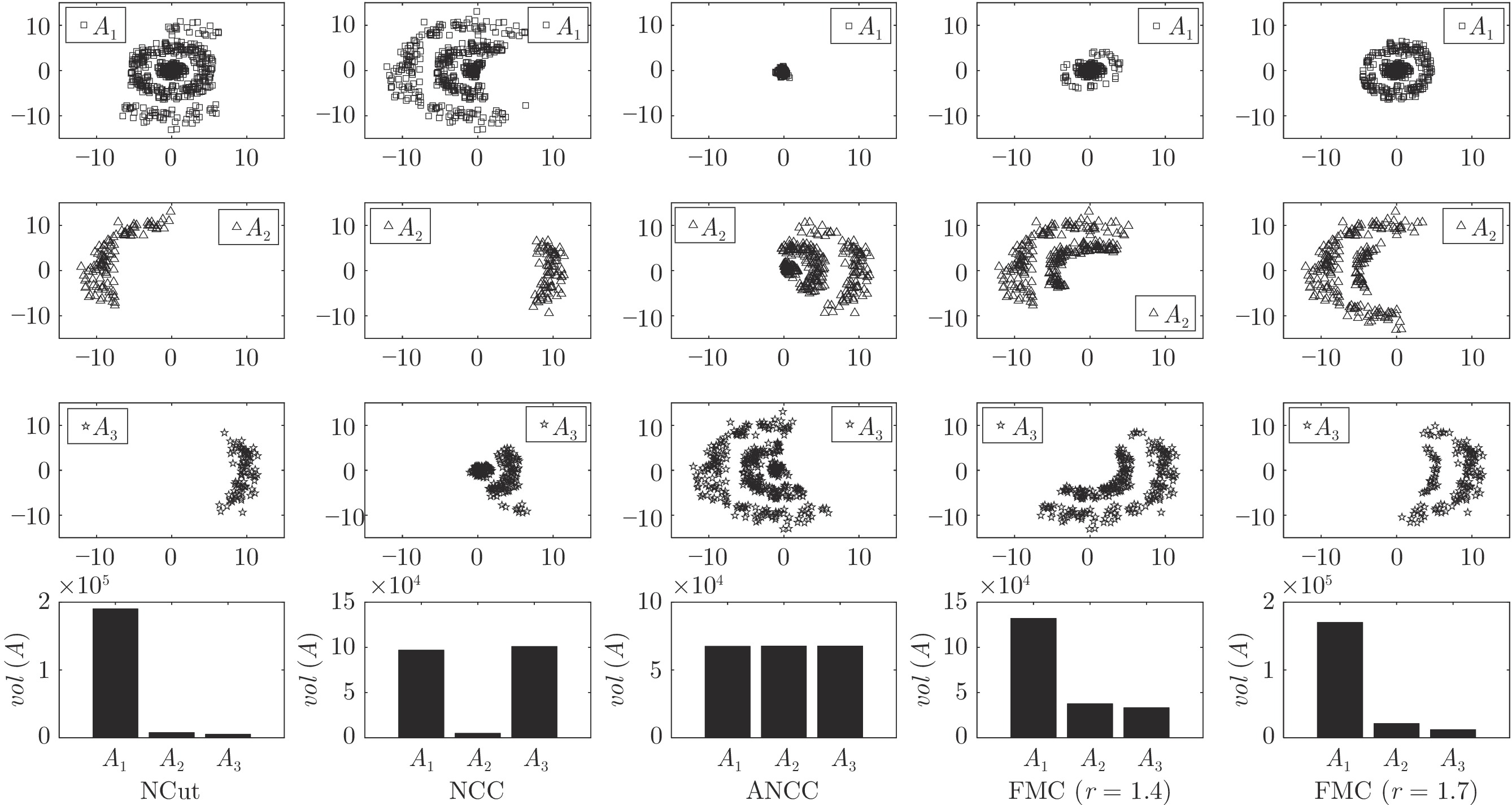
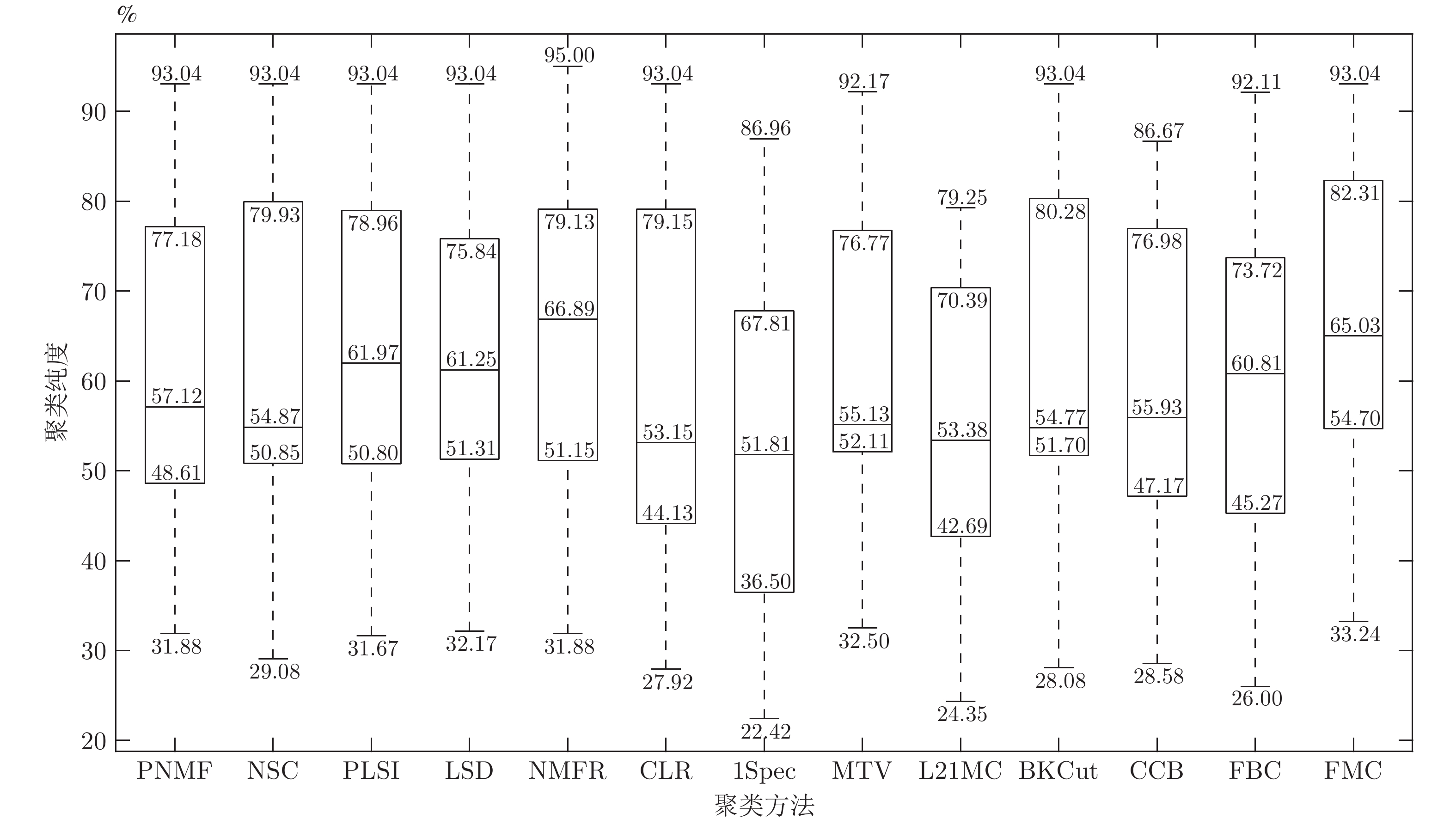
数据集 1Spec[16] MTV[11] L21MC[19] BKCut[18] CCB[15] FBC (r) FMC (r) HIGHSCHOOL 81.67 80.00 70.00 81.67 86.67 71.67 (0.9) 89.67 (1.2) POLBOOKS 82.86 79.05 77.14 81.90 83.81 81.50 (1.4) 83.81 (1.3) FOOTBALL 86.96 92.17 24.35 93.04 86.09 92.11 (1.3) 93.04 (1.8) DUKE 52.27 70.45 63.64 52.27 52.27 70.45 (0.5) 70.45 (0.5) CANCER 34.34 54.04 52.02 54.55 48.48 53.54 (1.5) 54.55 (1.7) KHAN 57.83 49.40 54.22 51.81 57.83 50.28 (1.6) 57.83 (1.6) ROSETTA 76.67 76.67 76.67 76.67 76.67 76.67 (1.0) 77.33 (1.9) ORL 36.00 52.50 79.25 82.00 74.25 74.75 (1.5) 81.75 (1.0) UMIST 58.96 76.87 70.78 70.09 65.22 72.70 (1.3) 74.96 (1.3) ALPHADIGS 36.11 51.71 46.08 47.72 45.87 36.84 (2.0) 49.00 (2.0) COIL20 45.00 81.46 50.97 78.89 77.29 81.81 (1.0) 82.92 (1.2) SAVEE 25.21 32.50 31.46 31.46 29.17 31.02 (1.1) 33.24 (1.3) CASIA 22.42 32.92 28.50 28.08 28.58 26.00 (0.1) 35.54 (0.4) IEMOCAP 54.03 54.00 54.00 54.03 54.03 54.03 (1.9) 55.21 (1.5) MED 44.72 53.92 52.76 54.99 53.53 50.09 (1.4) 55.00 (1.4) WEBKB4 39.39 56.22 39.30 51.60 39.56 46.14 (0.5) 59.60 (0.7) REUTERS 52.83 73.34 65.69 61.02 69.38 67.58 (2.0) 73.53 (1.5) YEAST 51.35 53.97 49.26 53.71 51.95 44.41 (1.6) 54.85 (1.6) FAULTS 36.89 40.08 37.92 41.11 39.52 31.72 (1.8) 42.76 (1.7) ECOLI 79.82 77.67 77.67 82.87 77.98 71.43 (1.8) 82.87 (1.8) 表 3 本文方法与其他非图聚类方法的聚类精度(%)对比
Table 3 Comparison of clustering accuracies (%) between ours and other non-graph clustering methods
数据集 PNMF[44] NSC[45] PLSI[42] LSD[46] NMFR[47] CLR[43] FMC (r) HIGHSCHOOL 81.67 83.33 83.33 81.67 95.00 80.00 89.67 (1.2) POLBOOKS 78.10 80.95 78.10 78.10 79.05 80.95 83.81 (1.3) FOOTBALL 93.04 93.04 93.04 93.04 93.04 93.04 93.04 (1.8) DUKE 52.27 52.27 70.45 70.45 70.45 52.27 70.45 (0.5) CANCER 53.53 53.03 53.53 53.03 51.52 54.04 54.55 (1.7) KHAN 60.24 55.42 55.42 51.81 50.60 51.81 57.83 (1.6) ROSETTA 76.67 76.67 76.67 76.67 76.67 77.00 77.33 (1.9) ORL 81.50 82.25 82.50 81.25 82.50 80.00 81.75 (1.0) UMIST 64.00 68.00 68.52 68.35 71.83 70.26 74.96 (1.3) ALPHADIGS 44.94 49.43 48.65 49.36 50.78 36.47 49.00 (2.0) COIL20 75.21 81.25 80.69 75.00 82.85 85.21 82.92 (1.2) SAVEE 31.88 31.46 31.67 34.17 31.88 28.13 33.24 (1.3) CASIA 35.17 29.08 32.00 32.17 34.08 27.92 35.54 (0.4) IEMOCAP 54.00 54.06 54.00 54.00 54.00 54.02 55.21 (1.5) MED 53.92 53.73 54.50 54.60 55.86 49.18 55.00 (1.4) WEBKB4 39.06 39.77 48.90 50.81 63.32 39.56 59.60 (0.7) REUTERS 73.86 76.47 75.92 74.99 76.52 51.80 73.53 (1.5) YEAST 52.76 54.31 52.69 52.09 54.85 46.90 54.85 (1.6) FAULTS 39.26 40.08 40.03 40.18 38.17 41.37 42.76 (1.7) ECOLI 77.68 78.90 79.82 67.89 79.20 78.29 82.87 (1.8) 表 4 不同聚类方法得到的图分割函数值
Table 4 The values of different graph cut functions obtained by different clustering methods
图分割函数 1Spec[16] MTV[11] L21MC[19] BKCut[18] CCB[15] PLSI[42] LSD[46] NMFR[47] CLR[43] 本文方法 FRCut0.5 12.64 (13.01) 13.99 (12.66) 15.57 (10.64) 12.03 (11.09) 18.63 (14.63) 14.43 (12.16) 14.70 (12.41) 14.27 (12.04) 13.69 (12.14) 9.71 (10.52) FRCut1.0 9.21 (9.64) 12.27 (11.22) 12.09 (8.27) 9.67 (9.31) 13.93 (11.28) 12.36 (10.35) 12.95 (10.83) 12.77 (10.90) 9.78 (9.08) 8.05 (8.18) FRCut2.0 7.79 (7.96) 11.89 (10.99) 10.90 (7.77) 8.37 (8.42) 12.18 (10.64) 11.88 (9.99) 12.62 (10.57) 12.50 (10.78) 8.18 (8.36) 6.72 (7.18) FNCut0.5 1.83 (1.48) 1.78 (1.42) 2.05 (1.25) 1.57 (1.25) 1.87 (1.46) 1.84 (1.35) 1.90 (1.38) 1.83 (1.37) 1.79 (1.48) 1.39 (1.21) FNCut1.0 1.35 (1.14) 1.53 (1.23) 1.56 (0.93) 1.29 (1.09) 1.41 (1.13) 1.54 (1.14) 1.63 (1.18) 1.60 (1.20) 1.26 (1.06) 1.14 (1.03) FNCut2.0 1.19 (1.00) 1.47 (1.20) 1.39 (0.87) 1.18 (1.03) 1.24 (1.07) 1.47 (1.10) 1.57 (1.14) 1.55 (1.17) 1.04 (0.95) 0.92 (0.83) -
[1] MacQueen J. Some methods for classification and analysis of multivariate observations. In: Proceedings of the 5th Berkeley Symposium on Mathematical Statistics and Probability. Berkeley, USA: University of California Press, 1967. 281−297 [2] Von Luxburg U. A tutorial on spectral clustering. Statistics and Computing, 2007, 17(4): 395-416 doi: 10.1007/s11222-007-9033-z [3] Ding S F, Cong L, Hu Q K, Jia H J, Shi Z Z. A multiway p-spectral clustering algorithm. Knowledge-Based Systems, 2019, 164: 371-377 doi: 10.1016/j.knosys.2018.11.007 [4] Liang N Y, Yang Z Y, Li Z N, Xie S L, Su C Y. Semi-supervised multi-view clustering with graph-regularized partially shared non-negative matrix factorization. Knowledge-Based Systems, 2020, 190: Article No. 105185 doi: 10.1016/j.knosys.2019.105185 [5] 庞宁, 张继福, 秦啸. 一种基于多属性权重的分类数据子空间聚类算法. 自动化学报, 2018, 44(3): 517-532Pang Ning, Zhang Ji-Fu, Qin Xiao. A subspace clustering algorithm of categorical data using multiple attribute weights. Acta Automatica Sinica, 2018, 44(3): 517-532 [6] 尹明, 吴浩杨, 谢胜利, 杨其宇. 基于自注意力对抗的深度子空间聚类. 自动化学报, 2022, 48(1): 271-281 doi: 10.16383/j.aas.c200302Yin Ming, Wu Hao-Yang, Xie Sheng-Li, Yang Qi-Yu. Self-attention adversarial based deep subspace clustering. Acta Automatica Sinica, 2022, 48(1): 271-281 doi: 10.16383/j.aas.c200302 [7] Stoer M, Wagner F. A simple min-cut algorithm. Journal of the ACM, 1997, 44(4): 585-591 doi: 10.1145/263867.263872 [8] Hagen L, Kahng A B. New spectral methods for ratio cut partitioning and clustering. IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems, 1992, 11(9): 1074-1085 doi: 10.1109/43.159993 [9] Shi J B, Malik J. Normalized cuts and image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2000, 22(8): 888-905 doi: 10.1109/34.868688 [10] Chung F R K. Spectral Graph Theory. Providence: American Mathematical Society, 1997. 36−45 [11] Bresson X, Laurent T, Uminsky D, Von Brecht J H. Multiclass total variation clustering. In: Proceedings of the 26th International Conference on Neural Information Processing Systems. Lake Tahoe, USA: Curran Associates Inc., 2013. 1421−1429 [12] Bresson X, Tai X C, Chan T F, Szlam A. Multi-class transductive learning based on ℓ1 relaxations of cheeger cut and Mumford-shah-Potts model. Journal of Mathematical Imaging and Vision, 2014, 49(1): 191-201 doi: 10.1007/s10851-013-0452-5 [13] Hein M, Setzer S. Beyond spectral clustering-tight relaxations of balanced graph cuts. In: Proceedings of the 24th International Conference on Neural Information Processing Systems. Granada, Spain: Curran Associates Inc., 2011. 2366−2374 [14] Bühler T, Hein M. Spectral clustering based on the graph p-laplacian. In: Proceedings of the 26th Annual International Conference on Machine Learning. Montreal, Canada: ACM, 2009. 81−88 [15] Cahill N D, Hayes T L, Meinhold R T, Hamilton J F. Compassionately conservative balanced cuts for image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. Salt Lake City, USA: IEEE, 2018. 1683−1691 [16] Hein M, Bühler T. An inverse power method for nonlinear eigenproblems with applications in 1-spectral clustering and sparse PCA. In: Proceedings of the 23rd International Conference on Neural Information Processing Systems. Vancouver, Canada: Curran Associates Inc., 2010. 847−855 [17] Nie F P, Wang H, Deng C, Gao X B, Li X L, Huang H. New ℓ1-norm relaxations and optimizations for graph clustering. In: Proceedings of the 30th AAAI Conference on Artificial Intelligence. Phoenix, USA: AAAI Press, 2016. 1962−1968 [18] Rangapuram S S, Mudrakarta P K, Hein M. Tight continuous relaxation of the balanced k-cut problem. In: Proceedings of the 27th International Conference on Neural Information Processing Systems. Montreal, Canada: MIT Press, 2014. 3131−3139 [19] Yang X, Deng C, Liu X L, Nie F P. New ℓ2,1-norm relaxation of multi-way graph cut for clustering. In: Proceedings of the 32nd AAAI Conference on Artificial Intelligence and the 30th Innovative Applications of Artificial Intelligence Conference and the 8th AAAI Symposium on Educational Advances in Artificial Intelligence. New Orleans, USA: AAAI Press, 2018. 4374−4381 [20] Ling S Y, Strohmer T. Certifying global optimality of graph cuts via semidefinite relaxation: A performance guarantee for spectral clustering. Foundations of Computational Mathematics, 2020, 20(3): 367-421 doi: 10.1007/s10208-019-09421-3 [21] Lovász L. Submodular functions and convexity. Mathematical Programming the State of the Art. Berlin, Heidelberg: Springer, 1983. 235−257 [22] Bach F. Submodular functions: From discrete to continuous domains. Mathematical Programming, 2019, 175(1-2): 419-459 doi: 10.1007/s10107-018-1248-6 [23] Ji S, Xu D C, Li M, Wang Y S, Zhang D M. Stochastic greedy algorithm is still good: Maximizing submodular + supermodular functions. In: Proceedings of the 6th World Congress on Global Optimization. Metz, France: Springer, 2019. 488−497 [24] Chambolle A, Pock T. A first-order primal-dual algorithm for convex problems with applications to imaging. Journal of Mathematical Imaging and Vision, 2011, 40(1): 120-145 doi: 10.1007/s10851-010-0251-1 [25] Batagelj V, Mrvar A. Pajek datasets [Online], available: http://vlado.fmf.uni-lj.si/pub/networks/data/, February 24, 2007 [26] Krebs V. Books about US politics [Online], available: https://www.cise.ufl.edu/research/sparse/matrices/Newman/polbooks.html, March 12, 2014 [27] Chang C C, Lin C J. LIBSVM — A library for support vector machines [Online], available: http://www.csie.ntu.edu.tw/~cjlin/libsvm, September 11, 2019 [28] Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York: Springer, 2009. 649−698 [29] Kim P M, Tidor B. Subsystem identification through dimensionality reduction of large-scale gene expression data. Genome Research, 2003, 13(7): 1706-1718 doi: 10.1101/gr.903503 [30] Samaria F S, Harter A C. Parameterisation of a stochastic model for human face identification. In: Proceedings of the IEEE Workshop on Applications of Computer Vision. Sarasota, USA: IEEE, 1994. 138−142. [31] Graham D B, Allinson N M. Characterising virtual eigensignatures for general purpose face recognition. Face Recognition: From Theory to Applications. Berlin: Springer, 1998. 446−456 [32] Roweis S. Binary alphadigits [Online], available: https://cs.nyu.edu/~roweis/data.html, January 12, 2010 [33] Nene S A, Nayar S K, Murase H. Columbia object image library (COIL-20) [Online], available: http://www.cs.columbia.edu/CAVE/software/softlib/coil-20.php, June 29, 2016 [34] Haq S, Jackson P J B, Edge J. Audio-visual feature selection and reduction for emotion classification. In: Proceedings of the International Conference on Auditory-Visual Speech Processing. Moreton Island, Australia: AVSP, 2008. 185−190 [35] 中文语言资源联盟. CASIA 汉语情感语料库 [Online], available: http://www.chineseldc.org/, 2010-10-09Chinese LDC. CASIA-Chinese emotional speech corpus [Online], available: http://www.chineseldc.org/, October 9, 2010 [36] Busso C, Bulut M, Lee C C, Kazemzadeh A, Mower E, Kim S, et al. IEMOCAP: Interactive emotional dyadic motion capture database. Language Resources and Evaluation, 2008, 42(4): 335-359 doi: 10.1007/s10579-008-9076-6 [37] Berry W M, Dumais S. LSI: Latent semantic indexing [Online], available: http://web.eecs.utk.edu/research/lsi/, November 21, 2008 [38] Srivastava S K, Singh S K, Suri J S. Effect of incremental feature enrichment on healthcare text classification system: A machine learning paradigm. Computer Methods and Programs in Biomedicine, 2019, 172: 35-51 doi: 10.1016/j.cmpb.2019.01.011 [39] Dua D, Graff C. UCI machine learning repository [Online], available: http://archive.ics.uci.edu/ml, September 24, 2018 [40] Oyallon E, Zagoruyko S, Huang G, Komodakis N, Julien S L, Blaschko M, et al. Scattering networks for hybrid representation learning. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2019, 41(9): 2208-2221 doi: 10.1109/TPAMI.2018.2855738 [41] Schuller B W, Batliner A, Bergler C, Messner E M, Hamilton A, Amiriparian S, et al. The INTERSPEECH 2020 computational paralinguistics challenge: Elderly emotion, breathing & masks. In: Proceedings of the 21st Annual Conference of the International Speech Communication Association. Shanghai, China: ISCA, 2020. 2042−2046 [42] Zhou D X, Yang D, Zhang X H, Huang S, Feng S. Discriminative probabilistic latent semantic analysis with application to single sample face recognition. Neural Processing Letters, 2019, 49(3): 1273-1298 doi: 10.1007/s11063-018-9852-2 [43] Nie F P, Wang X Q, Jordan M I, Huang H. The constrained Laplacian rank algorithm for graph-based clustering. In: Proceedings of the 30th AAAI Conference on Artificial Intelligence. Phoenix, USA: AAAI Press, 2016. 1969−1976 [44] Yang Z R, Oja E. Linear and nonlinear projective nonnegative matrix factorization. IEEE Transactions on Neural Networks, 2010, 21(5): 734-749 doi: 10.1109/TNN.2010.2041361 [45] Ding C, Li T, Jordan M I. Nonnegative matrix factorization for combinatorial optimization: Spectral clustering, graph matching, and clique finding. In: Proceedings of the 8th IEEE International Conference on Data Mining. Pisa, Italy: IEEE, 2008. 183−192 [46] Arora R, Gupta M R, Kapila A, Fazel M. Clustering by left-stochastic matrix factorization. In: Proceedings of the 28th International Conference on International Conference on Machine Learning. Bellevue, USA: Omnipress, 2011. 761−768 [47] Yang Z R, Hao T L, Dikmen O, Chen X, Oja E. Clustering by nonnegative matrix factorization using graph random walk. In: Proceedings of the 25th International Conference on Neural Information Processing Systems. Lake Tahoe, USA: Curran Associates Inc., 2012. 1079−1087 -








 下载:
下载: