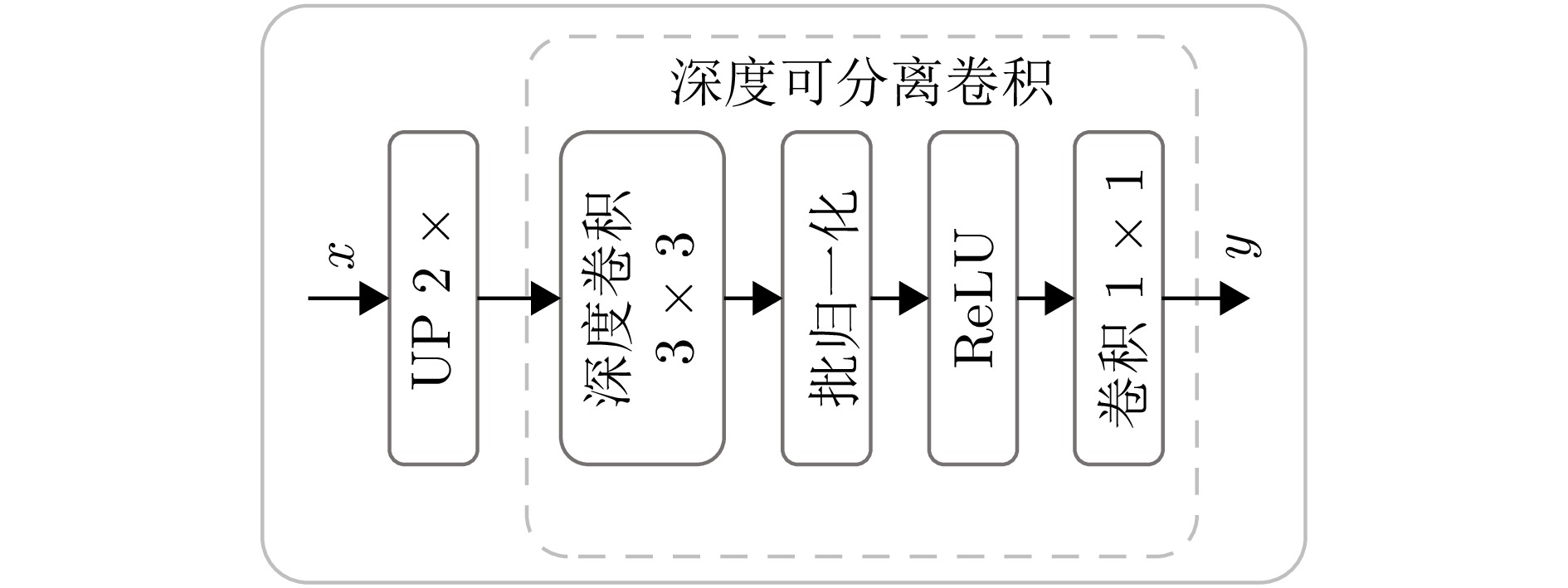
-
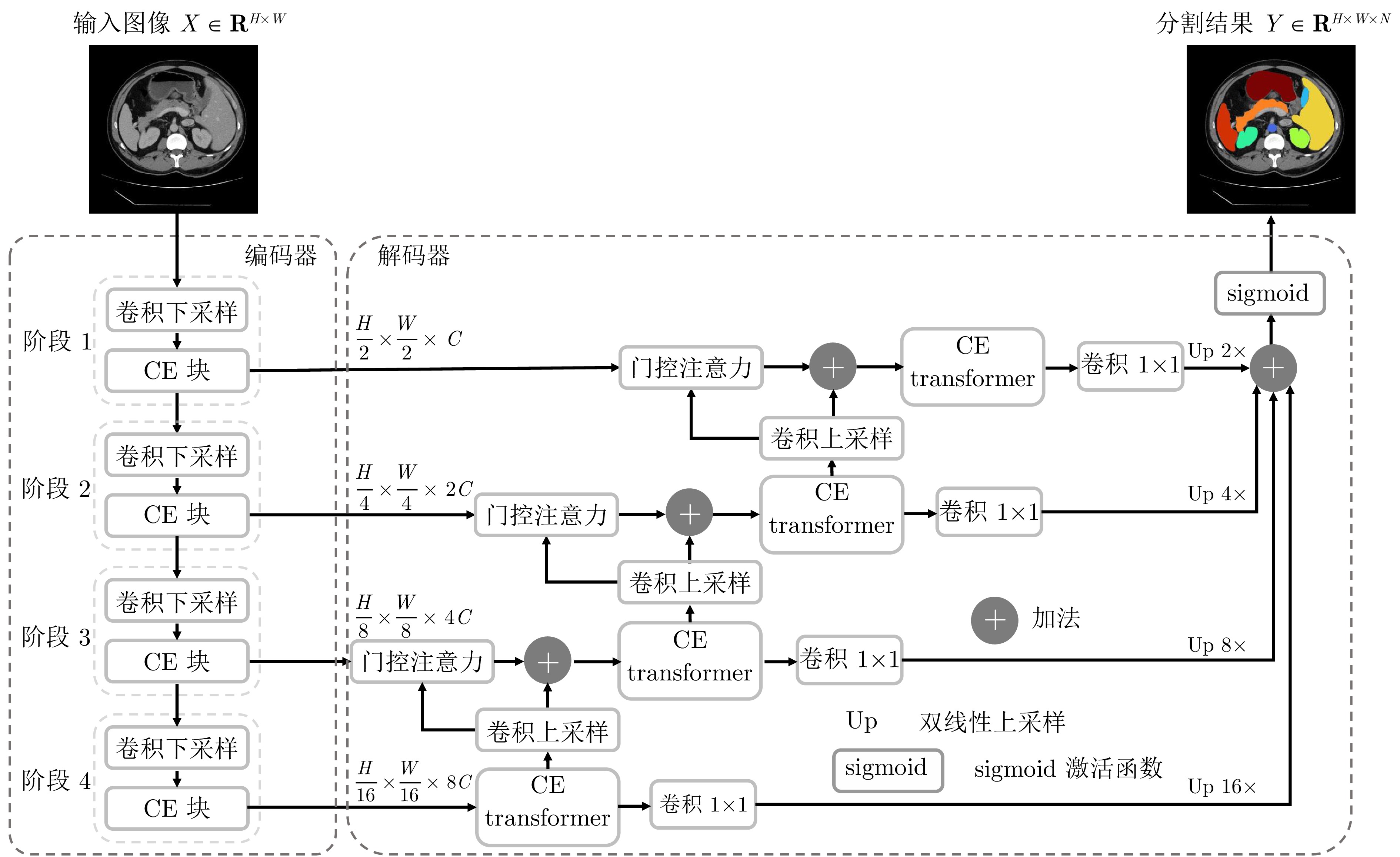
摘要: 受限于局部感受野, 卷积神经网络无法建立足够的长距离依赖关系. 一些方法通过将Transformer部署至卷积网络的某些特定部位来缓解这个问题, 如网络编码器、解码器或跳跃连接层. 但这些方法只能为某些特定特征建立长距离依赖关系, 难以捕获大小、形态多样的腹部器官之间的复杂依赖关系. 针对该问题, 提出一种交叉增强Transformer (CE transformer)结构, 并用它作为特征提取单元构建一种新的多层级编−解码分割网络CE TransNet. CE transformer采用双路径设计, 深度融合Transformer与卷积结构, 可同时对长、短距离依赖关系进行建模. 在双路径中, 引入密集交叉连接促进不同粒度信息的交互与融合, 提高模型整体特征捕获能力. 将CE transformer部署于CE TransNet的整个编解码路径中, 可有效捕捉多器官的复杂上下文关系. 实验结果表明, 所提出方法在WORD和Synapse腹部CT多器官数据集上的平均Dice相似系数值分别高达82.42%和81.94%, 显著高于多种当前先进方法.
-
关键词:
- 多器官分割 /
- 深度学习 /
- Transformer /
- 交叉连接
Abstract: Limited by local receptive fields, convolutional neural network is unable to establish sufficient long-range dependencies. Some methods address this limitation by deploying transformers to some specific parts of the convolutional network, such as the encoder, decoder, or skip connections. However, these methods can only establish long-range dependencies for certain specific features and struggle to capture the intricate dependencies among diverse-sized and shaped abdominal organs. To tackle this issue, a cross-connection enhanced transformer (CE transformer) architecture is proposed. Utilizing the CE transformer as the feature extraction module, a multi-level encoder-decoder segmentation network, denoted as CE TransNet, is developed. The CE transformer allows simultaneous modeling of long- and short-range dependencies by employing a dual-path design that integrates Transformer and convolutional structures. Dense cross-connections are introduced in the dual paths to facilitate the interaction and fusion of information in different granularity, enhancing the overall feature capture capability of the model. The CE transformer deployed across the entire encoding and decoding paths of the CE TransNet, which allows for establishing complex dependencies of organs. Experimental results show that the proposed method achieves mean Dice similarity coefficient values of 82.42% and 81.94% on the WORD and Synapse abdominal CT multi-organ datasets, respectively, which are significantly higher than those of many state-of-the-art methods.-
Key words:
- Multi-organ segmentation /
- deep learning /
- Transformer /
- cross connection
-
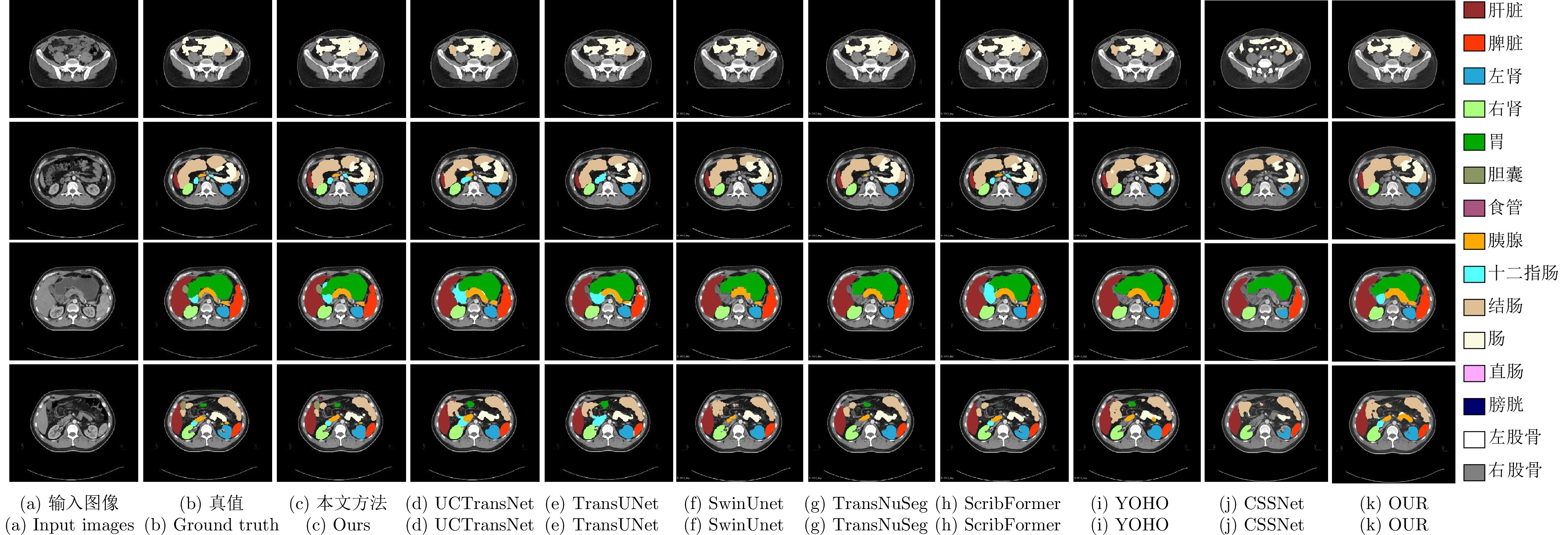
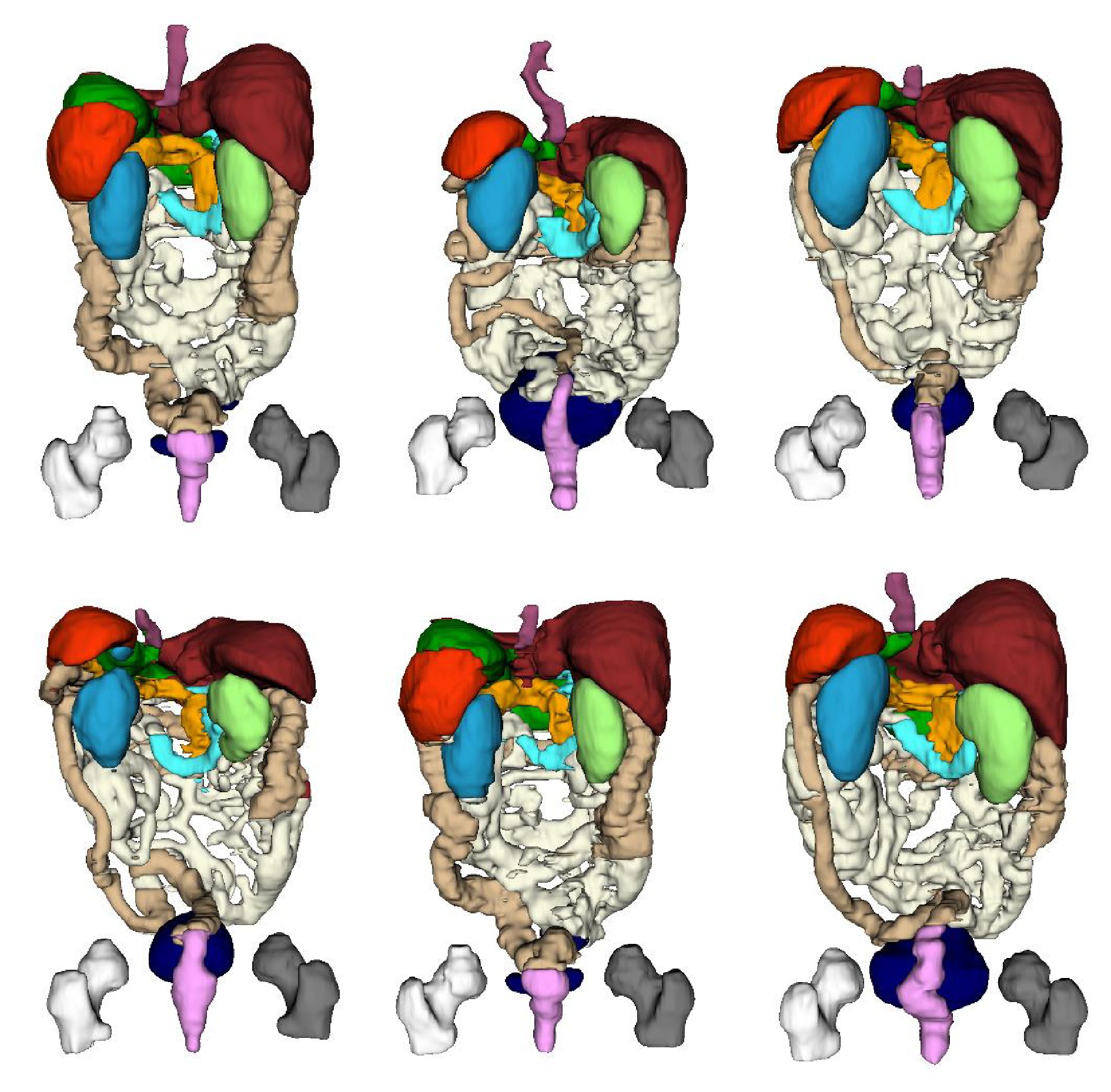
表 1 不同方法在WORD数据集上的平均分割性能比较
Table 1 Average segmentation performance comparison of different methods on the WORD dataset
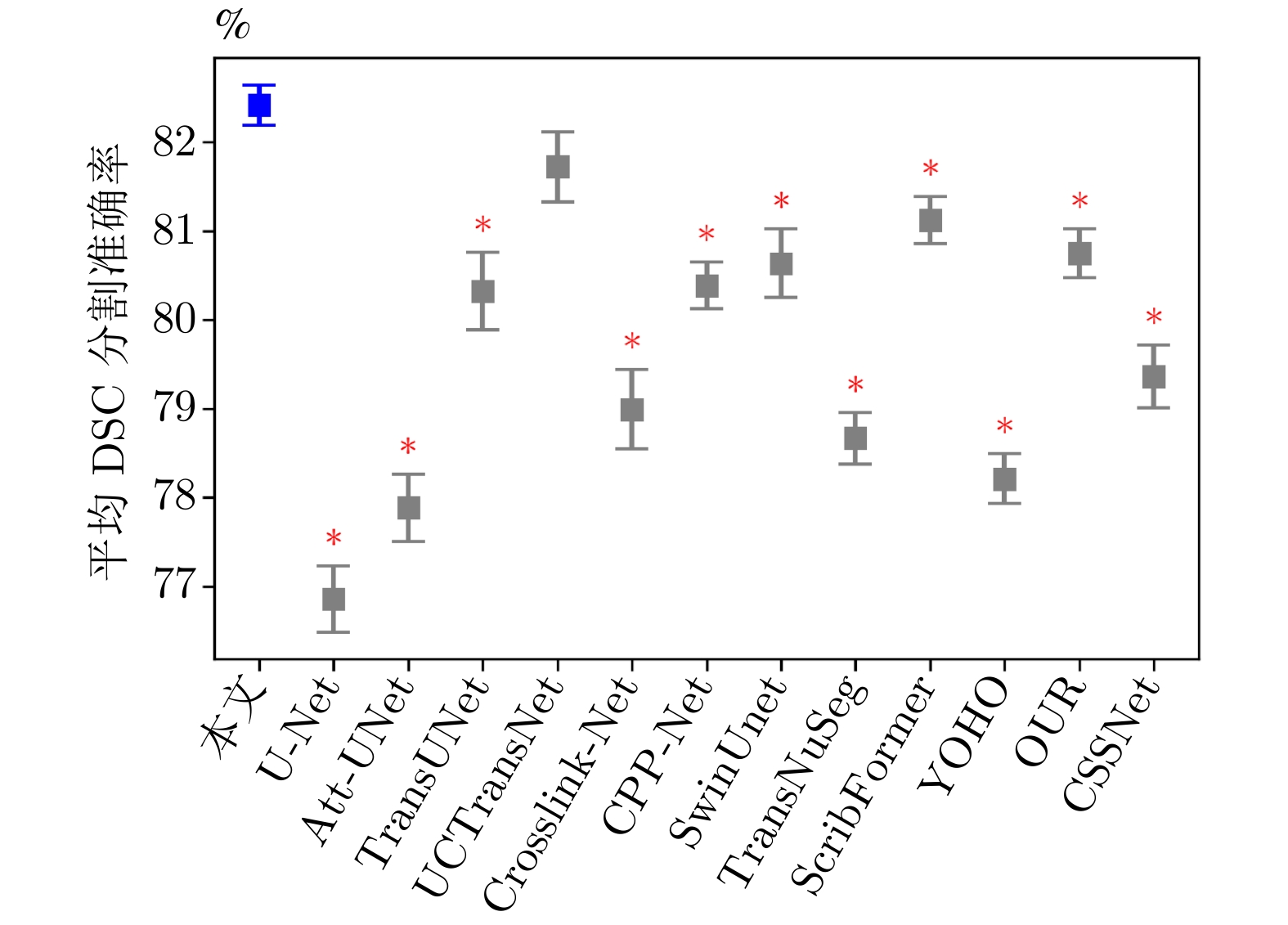
方法 出版机构/年份 DSC (%)$ \uparrow $ mIoU (%)$ \uparrow $ NSD (%)$ \uparrow $ HD (mm)$ \downarrow $ ASD (mm)$ \downarrow $ Recall (%)$ \uparrow $ Precision (%)$ \uparrow $ U-Net[10] MICCAI/2015 76.93 65.35 62.03 17.16 4.44 85.13 78.53 Att-UNet[41] Elsevier MIA/2019 77.83 66.74 65.41 16.43 3.91 84.05 83.86 TransUNet[23] arXiv/2021 80.32 69.95 69.29 20.31 5.51 87.98 80.92 UCTransNet[25] AAAI/2022 81.64 71.34 69.78 11.30 2.67 86.10 84.16 Crosslink-Net[42] IEEE TIP/2022 78.99 68.15 65.33 13.13 2.88 81.62 83.10 CPP-Net[43] IEEE TIP/2023 80.36 70.04 70.76 12.82 2.98 85.31 84.53 SwinUnet[28] ECCV/2022 80.64 69.82 69.09 15.23 4.12 82.93 80.40 TransNuSeg[44] MICCAI/2023 78.63 68.31 67.73 14.41 2.89 85.78 80.06 ScribFormer[31] IEEE TMI/2024 81.21 71.07 73.08 11.78 2.91 85.34 84.43 YOHO[45] IEEE TIP/2024 78.23 67.45 65.67 13.68 3.29 81.86 81.97 OUR[46] MBEC/2023 80.71 70.06 71.38 12.06 2.92 87.38 84.77 CSSNet[47] CMB/2024 79.41 69.02 67.29 14.69 3.16 86.38 80.32 本文方法 — 82.42 72.48 74.34 10.91 2.62 86.47 85.35 注: 加粗字体表示不同方法在各指标上的最优结果. 表 3 不同方法在WORD数据集上的NSD得分比较 (%)
Table 3 NSD score comparison of different methods on the WORD dataset (%)
方法 肝脏 脾脏 左肾 右肾 胃 胆囊 食管 胰腺 十二指肠 结肠 肠 直肠 膀胱 左股骨 右股骨 U-Net[10] 74.13 70.24 68.66 69.66 57.09 47.43 50.99 58.93 41.21 55.63 60.85 54.86 72.70 72.10 75.94 Att-UNet[41] 81.01 82.45 74.52 77.72 64.73 42.45 66.92 61.37 44.56 57.80 63.49 53.67 70.75 65.90 73.88 TransUNet[23] 82.36 86.55 81.23 81.54 71.67 36.12 69.86 65.23 23.71 62.06 67.15 60.18 81.97 84.80 84.97 UCTransNet[25] 89.27 86.52 84.85 77.41 73.77 50.38 58.74 68.63 30.81 64.45 54.61 61.15 76.79 85.06 84.27 Crosslink-Net[42] 79.65 84.02 79.95 80.52 65.09 35.67 66.88 31.55 38.25 60.12 63.82 56.32 77.13 79.66 81.33 CPP-Net[43] 81.24 83.77 80.16 81.42 71.85 48.97 70.80 60.59 45.19 63.48 67.21 59.60 79.79 83.88 83.44 SwinUnet[28] 83.13 85.79 81.83 73.96 61.29 43.86 68.50 68.00 49.58 63.71 58.25 60.64 78.30 79.42 80.33 TransNuSeg[44] 79.78 77.37 78.18 78.50 60.65 45.15 70.96 68.35 47.78 61.69 60.98 58.89 75.99 75.23 76.48 ScribFormer[31] 82.34 84.19 83.19 81.51 70.14 47.88 71.31 66.49 48.51 63.59 66.86 60.25 80.02 84.43 84.86 YOHO[45] 77.14 80.16 76.23 77.75 62.88 49.13 61.62 53.60 48.57 53.57 59.26 53.01 74.21 79.30 78.65 OUR[46] 82.65 84.55 82.21 81.81 70.07 47.57 70.16 65.88 48.51 64.50 66.18 60.02 79.61 82.69 84.32 CSSNet[47] 81.33 82.83 80.45 80.41 68.19 45.70 68.86 63.69 47.74 62.65 64.10 59.18 75.00 78.67 76.56 本文方法 84.02 86.70 82.12 82.91 73.93 64.38 70.64 69.14 50.87 65.14 68.12 61.97 82.34 86.50 86.29 注: 加粗字体表示不同方法在各器官上的最优结果. 表 2 不同方法在WORD数据集上的DSC得分比较 (%)
Table 2 DSC score comparison of different methods on the WORD dataset (%)
方法 肝脏 脾脏 左肾 右肾 胃 胆囊 食管 胰腺 十二指肠 结肠 肠 直肠 膀胱 左股骨 右股骨 U-Net[10] 93.09 88.05 87.79 89.17 79.27 52.63 59.25 70.05 52.96 71.62 76.44 71.59 85.54 87.53 88.99 Att-UNet[41] 94.67 92.28 88.09 89.35 82.43 43.53 70.12 69.32 54.90 74.11 77.80 72.10 85.16 85.28 88.33 TransUNet[23] 94.80 93.19 90.09 90.25 87.04 48.30 72.66 73.26 52.21 77.11 80.00 74.40 89.31 90.97 91.24 UCTransNet[25] 94.63 92.58 90.19 89.38 87.34 60.78 73.95 75.09 59.33 76.74 78.71 74.60 89.07 91.00 91.15 Crosslink-Net[42] 94.52 92.98 90.81 91.53 83.89 56.44 70.71 65.15 48.00 75.21 77.44 72.47 85.38 89.80 90.56 CPP-Net[43] 94.56 92.35 89.75 90.59 87.42 50.22 73.01 70.36 56.75 77.61 79.83 73.28 88.37 90.55 90.74 SwinUnet[28] 94.92 93.99 89.26 90.82 85.17 56.71 68.02 72.48 57.54 75.56 79.55 74.81 88.86 90.46 91.40 TransNuSeg[44] 94.78 92.96 89.34 87.80 79.58 53.36 69.04 71.44 57.37 75.60 76.77 76.28 86.55 83.86 84.77 ScribFormer[31] 95.00 93.23 90.98 90.11 86.62 57.52 71.29 73.69 56.66 77.86 80.24 74.14 88.84 90.79 91.20 YOHO[45] 93.92 91.85 89.59 90.13 83.20 51.03 67.60 65.19 58.06 71.35 74.60 71.22 86.08 89.71 89.89 OUR[46] 95.08 93.60 90.28 90.10 86.45 58.92 68.66 73.41 55.08 76.86 78.53 73.62 88.52 90.41 91.22 CSSNet[47] 94.96 92.82 88.92 87.53 82.70 55.76 67.45 73.10 54.16 76.63 77.77 72.00 87.68 89.60 90.06 本文方法 95.43 94.20 91.26 91.67 87.08 63.52 72.14 75.80 59.89 78.05 80.83 74.93 88.39 91.43 91.75 注: 加粗字体表示不同方法在各器官上的最优结果. 表 4 不同方法在Synapse数据集上的分割性能比较
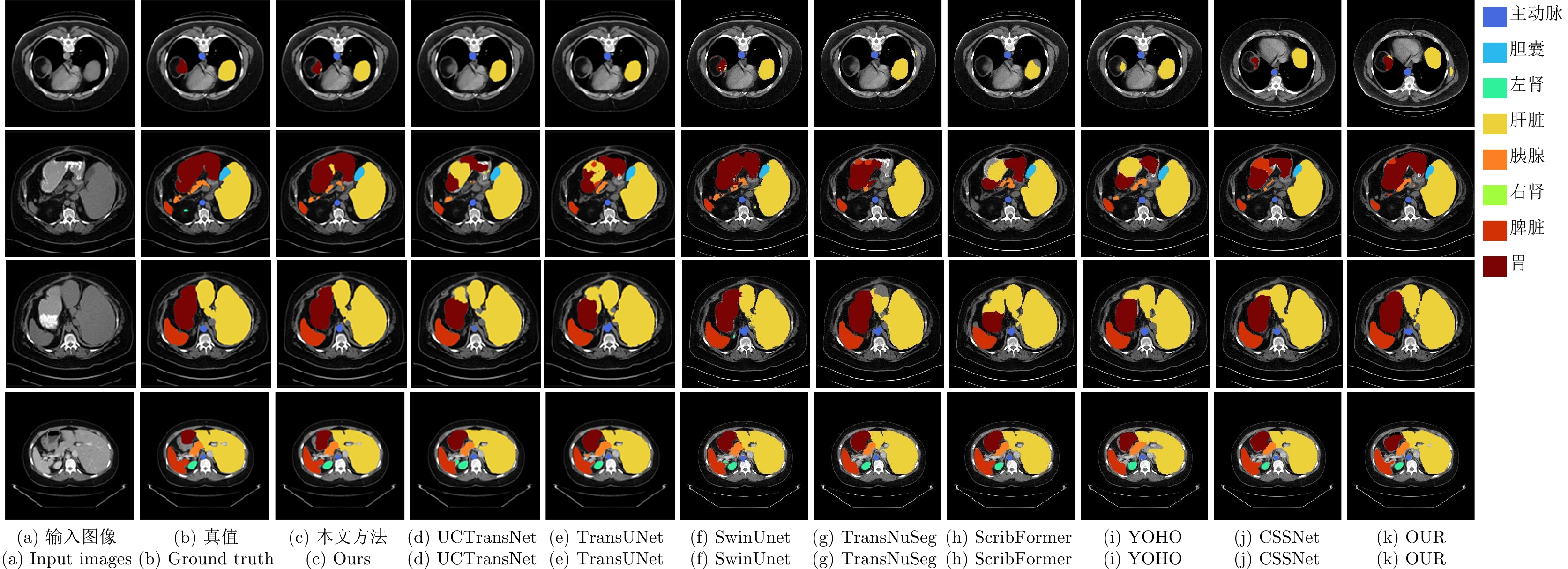
Table 4 Segmentation performance comparison of different methods on the Synapse dataset
方法 出版/年份 平均 各器官DSC (%) DSC (%)$ \uparrow $ HD (mm)$ \downarrow $ mIoU (%)$ \uparrow $ 主动脉 胆囊 左肾 右肾 肝脏 胰腺 脾脏 胃 U-Net[10] MICCAI/2015 70.11 44.69 59.39 84.00 56.70 72.41 62.64 86.98 48.73 81.48 67.96 Att-UNet[41] Elsevier MIA/2019 71.70 34.47 61.38 82.61 61.94 76.07 70.42 87.54 46.70 80.67 67.66 TransUNet[23] arXiv/2021 77.62 26.90 67.32 86.56 60.43 80.54 78.53 94.33 58.47 87.06 75.00 UCTransNet[25] AAAI/2022 80.21 23.33 70.46 87.36 66.49 83.77 79.95 94.23 63.72 89.38 76.75 Crosslink-Net[42] IEEE TIP/2022 76.60 18.20 64.83 86.25 53.35 84.62 79.63 92.72 58.56 86.17 71.49 CPP-Net[43] IEEE TIP/2023 80.11 26.41 71.23 87.59 67.14 83.09 82.31 94.03 67.34 87.53 71.81 SwinUnet[28] ECCV/2022 79.13 21.55 68.81 85.47 66.53 83.28 79.61 94.29 56.58 90.66 76.60 TransNuSeg[44] MICCAI/2023 78.06 28.69 69.03 82.47 65.94 79.05 79.11 93.12 58.40 88.85 77.49 ScribFormer[31] IEEE TMI/2024 80.08 20.78 70.63 87.48 65.15 86.90 82.09 94.26 60.48 88.93 75.37 YOHO[45] IEEE TIP/2024 76.85 27.41 67.79 85.34 66.33 83.38 73.66 93.82 55.57 82.65 74.07 OUR[46] MBEC/2023 80.06 27.54 69.72 88.32 65.96 87.02 82.50 94.31 60.23 88.41 73.76 CSSNet[47] CMB/2024 78.75 29.81 68.01 86.80 64.12 82.54 79.04 94.05 58.98 89.47 75.04 本文方法 — 81.94 22.54 71.42 89.79 67.97 88.54 84.12 94.56 63.09 91.56 75.90 注: 加粗字体表示不同方法在各指标上的最优结果. 表 5 不同特征提取模块的分割性能比较
Table 5 Segmentation performance comparison on different feature extraction module
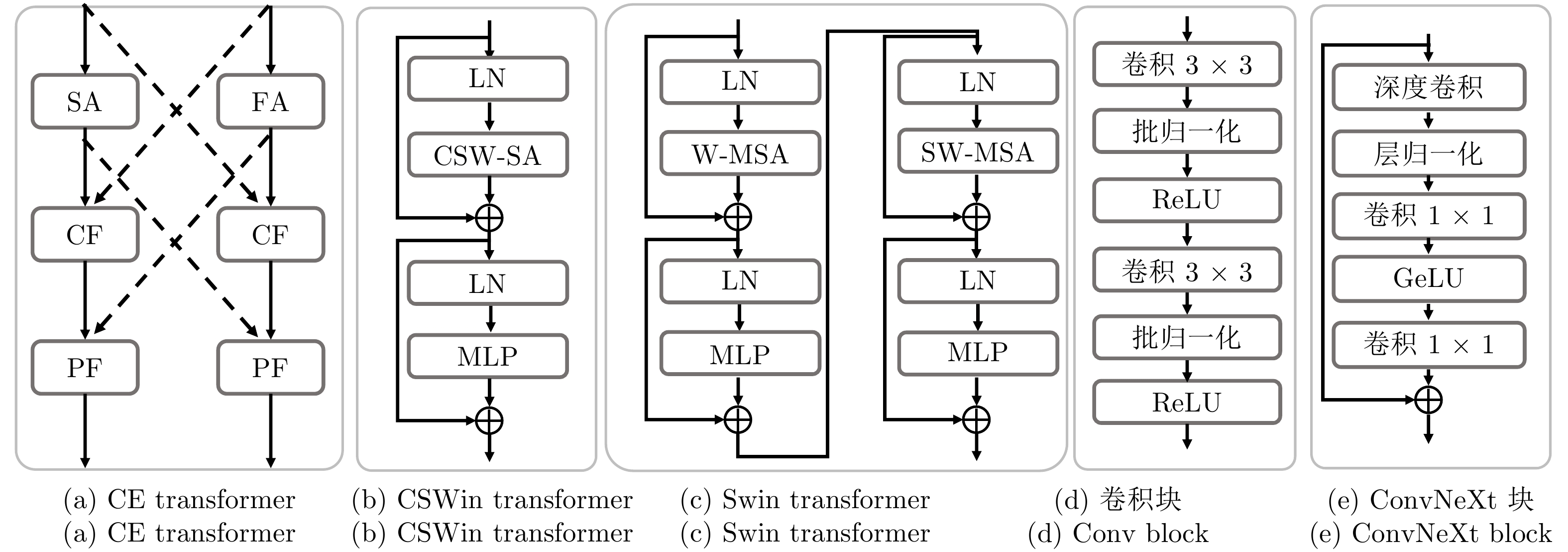
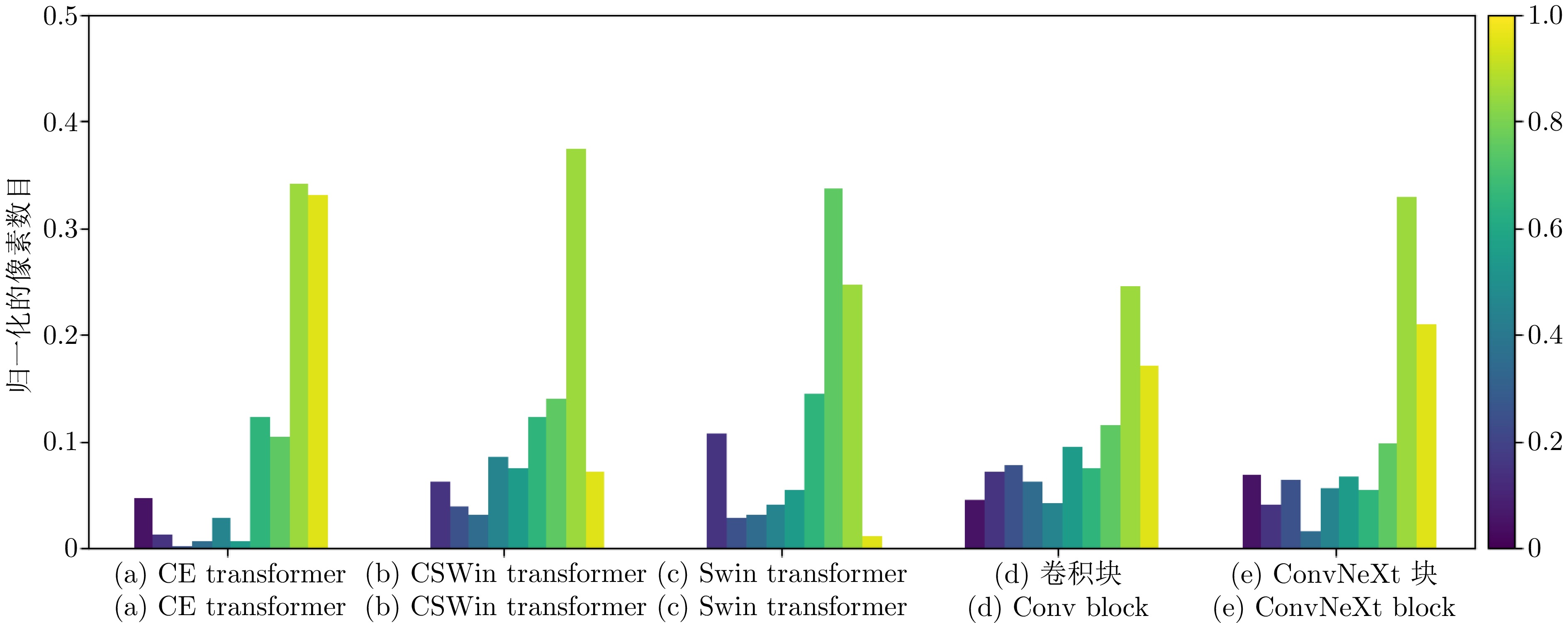
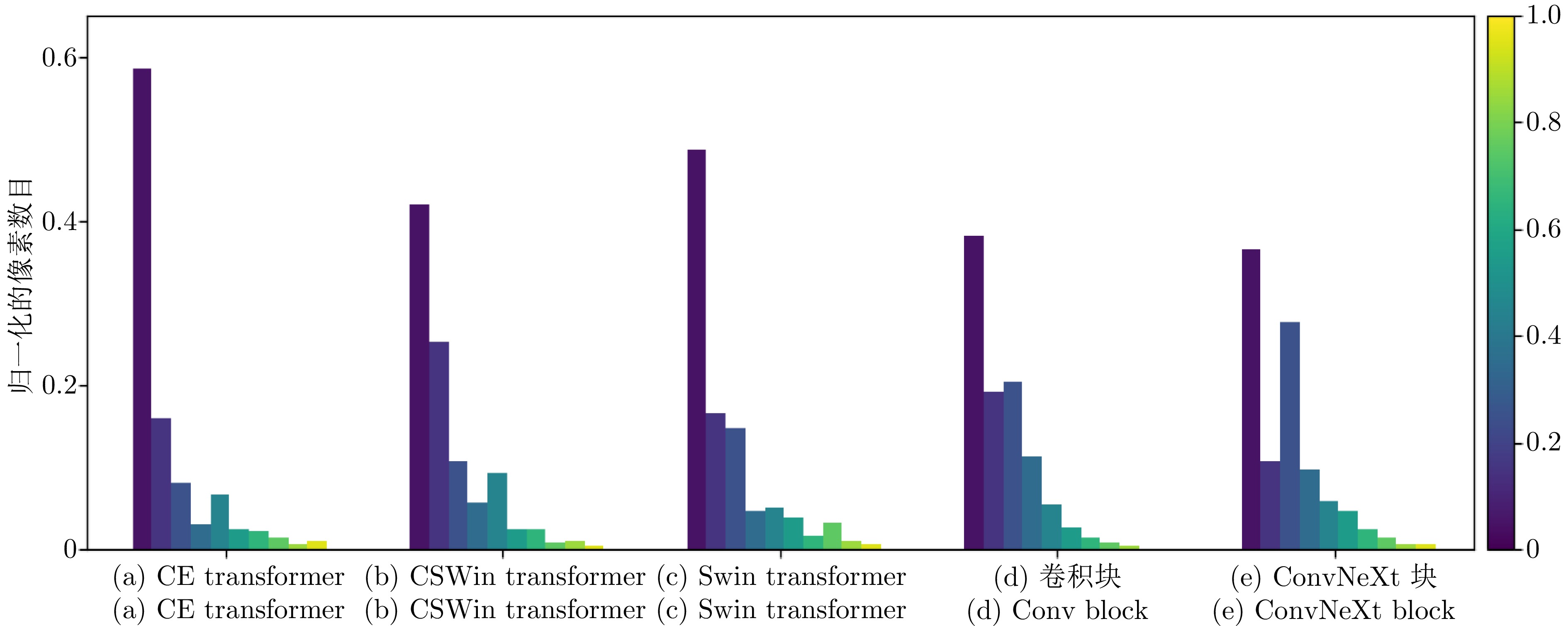
特征提取模块 FLOPS
(G)$ \downarrow $Params
(M)$ \downarrow $DSC
(%)$ \uparrow $HD
(mm)$ \downarrow $CE transformer 6.54 6.23 81.94 22.54 Swin transformer[35] 11.28 15.84 79.97 27.42 CSWin transformer[36] 11.29 15.87 80.33 24.31 卷积块 15.26 22.84 79.51 28.01 ConvNeXt块[51] 8.75 11.30 80.48 23.39 注: 加粗字体表示在各指标上的最优结果. 表 6 采用不同$ N $值的分割性能比较
Table 6 Segmentation performance comparison of applying different $ N $
N (阶段) FLOPS (G)$ \downarrow $ Params (M)$ \downarrow $ DSC (%)$ \uparrow $ HD (mm)$ \downarrow $ 1 2 3 4 0 0 0 0 4.17 3.16 73.26 43.46 1 1 1 1 5.16 4.59 77.43 35.78 2 2 2 2 6.12 6.02 79.52 24.22 1 2 4 2 6.54 6.23 81.94 22.54 1 3 6 3 7.15 8.22 81.33 24.89 2 4 8 4 8.33 9.92 81.43 25.42 2 6 12 6 10.10 13.28 81.67 23.11 注: 加粗字体表示在各指标上的最优结果. 表 7 不同交叉连接策略的分割性能比较
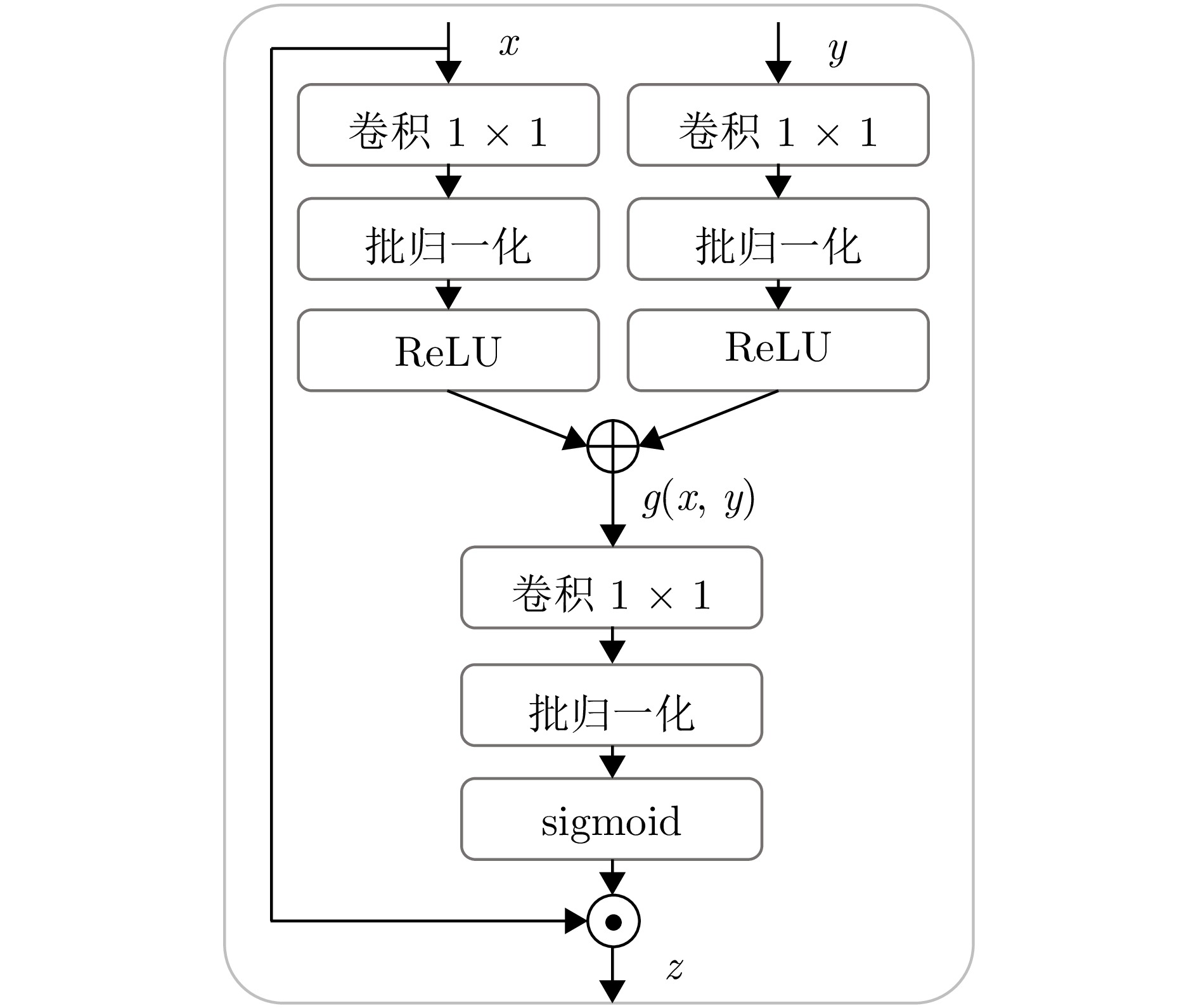
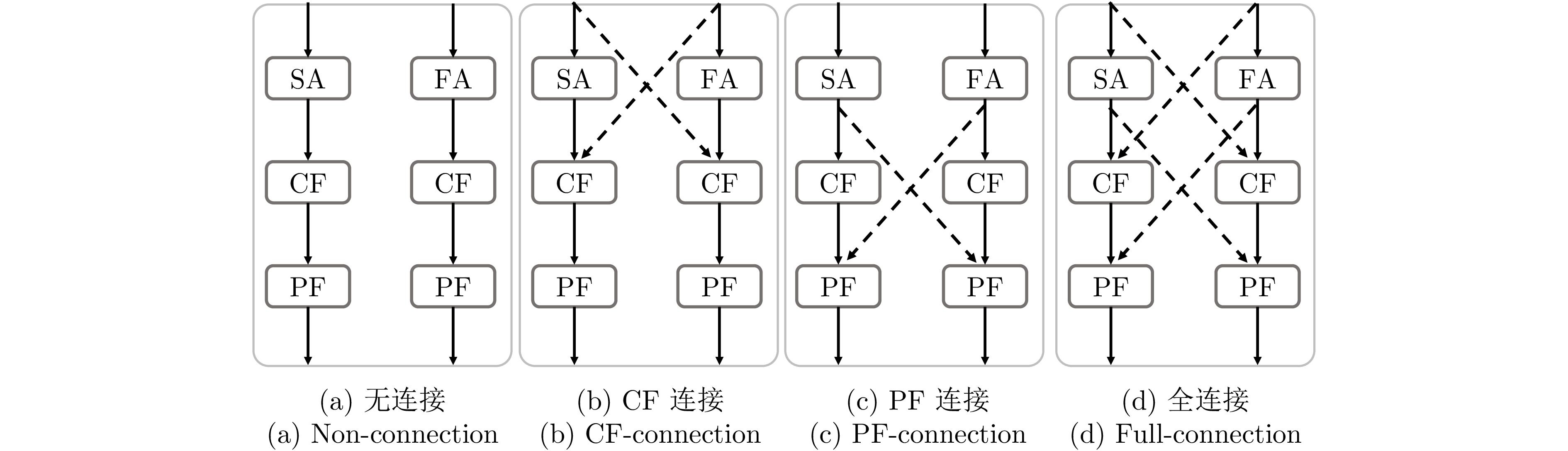
Table 7 Segmentation performance comparison of different cross-connection configurations
交叉连接策略 DSC (%)$ \uparrow $ HD (mm)$ \downarrow $ mIoU (%)$ \uparrow $ 无连接 79.97 27.81 70.04 CF连接 80.87 23.07 70.83 PF连接 80.19 24.28 70.76 全连接 81.94 22.54 71.42 注: 加粗字体表示不同方法在各指标上的最优结果. 表 8 CE transformer中各子模块对网络性能的影响
Table 8 Impact of each sub-module in CE transformer on network performance
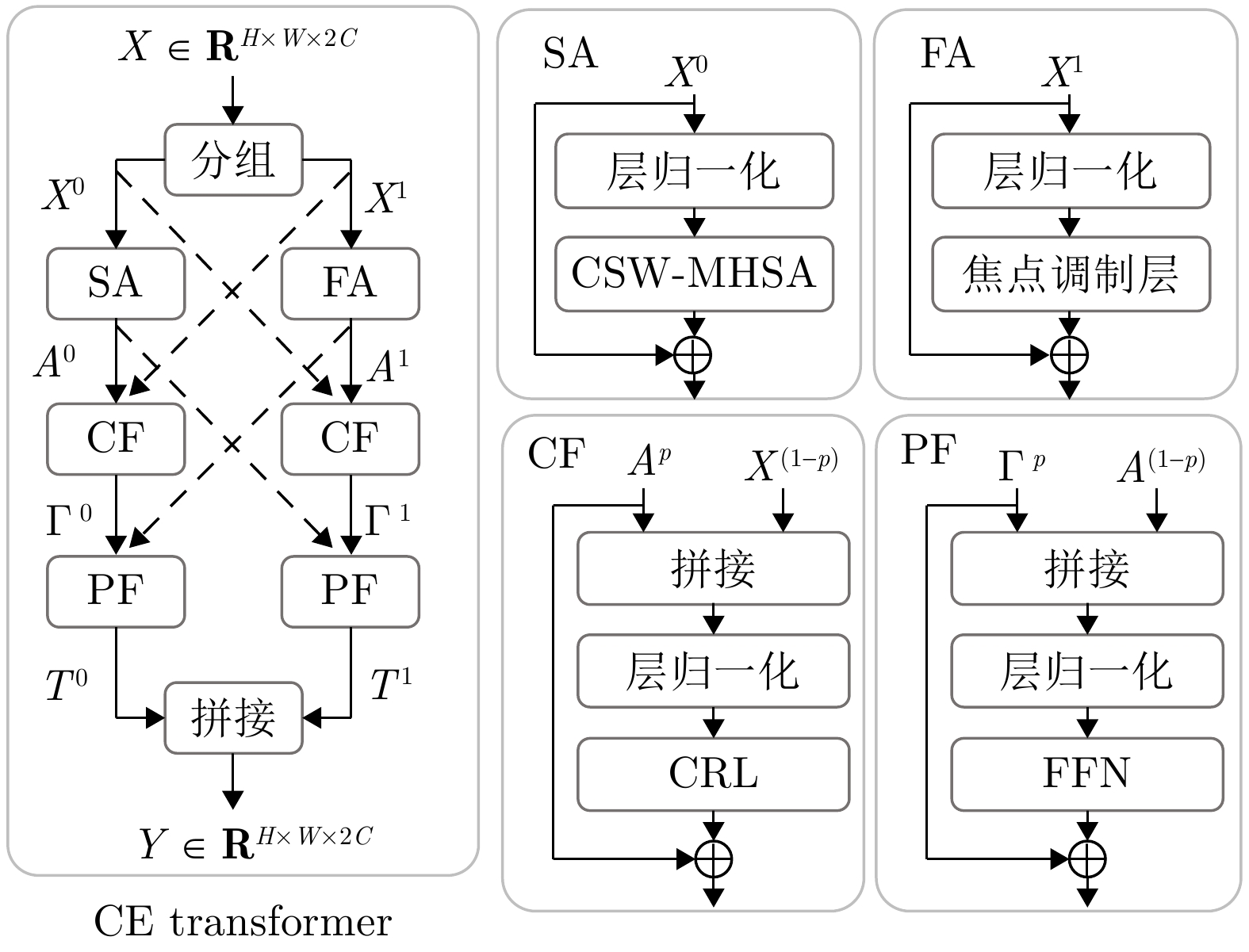
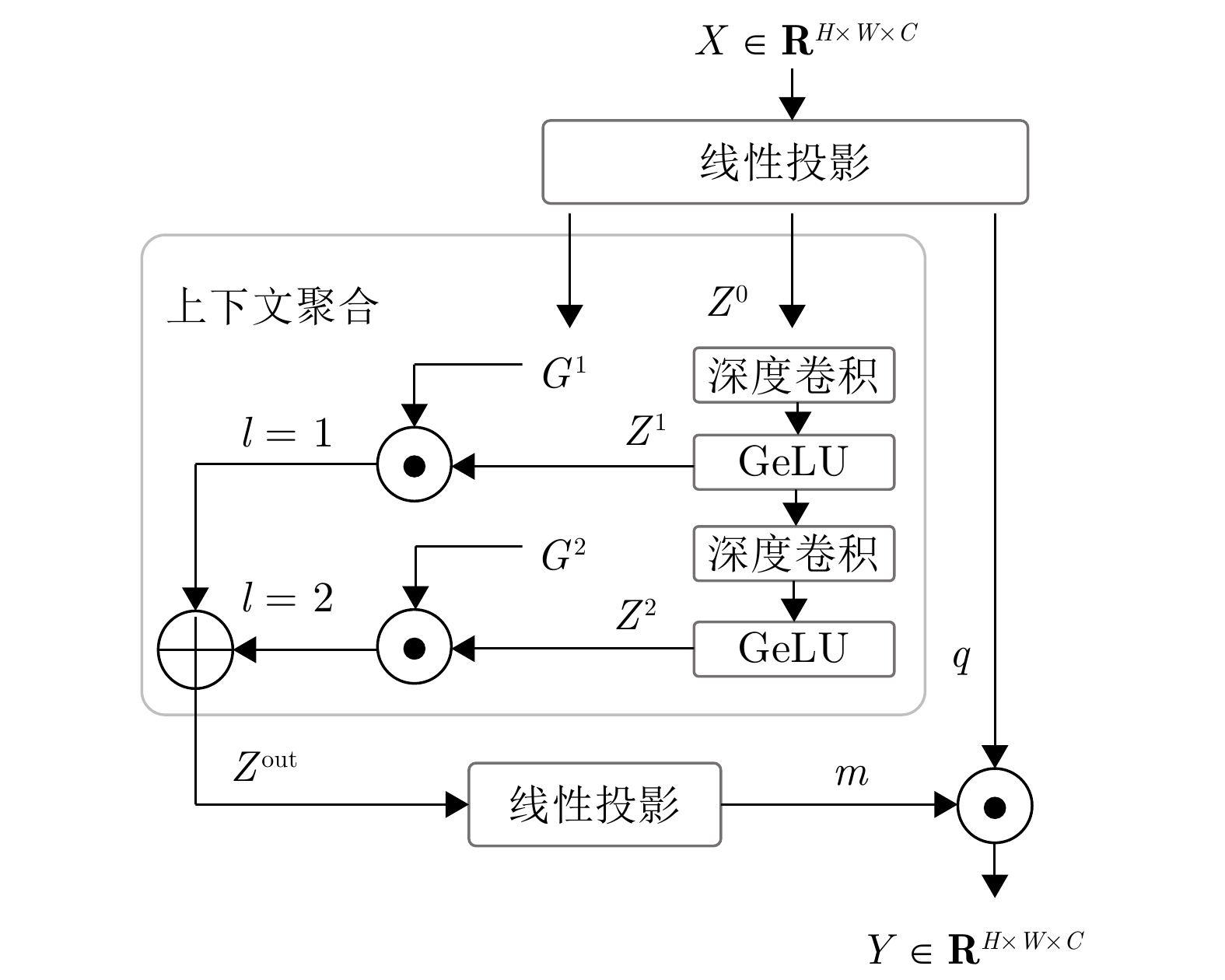
实验序号 SA FA CF PF DSC (%)$ \uparrow $ HD (mm)$ \downarrow $ 1 $ \checkmark$ 74.59 40.70 2 $ \checkmark$ $ \checkmark$ 78.48 26.13 3 $ \checkmark$ $ \checkmark$ $ \checkmark$ 80.31 23.31 4 $ \checkmark$ $ \checkmark$ $ \checkmark$ $ \checkmark$ 81.94 22.54 注: 加粗字体表示不同方法在各指标上的最优结果. -
[1] 方超伟, 李雪, 李钟毓, 焦李成, 张鼎文. 基于双模型交互学习的半监督医学图像分割. 自动化学报, 2023, 49(4): 805−819Fang Chao-Wei, Li Xue, Li Zhong-Yu, Jiao Li-Cheng, Zhang Ding-Wen. Interactive dual-model learning for semi-supervised medical image segmentation. Acta Automatica Sinica, 2023, 49(4): 805−819 [2] Ji Y F, Bai H T, Ge C J, Yang J, Zhu Y, Zhang R M, et al. AMOS: A large-scale abdominal multi-organ benchmark for versatile medical image segmentation. In: Proceedings of the 36th International Conference on Neural Information Processing Systems. New Orleans, USA: Curran Associates Inc., 2019. Article No. 2661 [3] Ma J, Zhang Y, Gu S, Zhu C, Ge C, Zhang Y C, et al. AbdomenCT-1K: Is abdominal organ segmentation a solved problem? IEEE Transactions on Pattern Analysis and Machine Intelligence, 2022, 44(10): 6695−6714 doi: 10.1109/TPAMI.2021.3100536 [4] 毕秀丽, 陆猛, 肖斌, 李伟生. 基于双解码U型卷积神经网络的胰腺分割. 软件学报, 2022, 33(5): 1947−1958Bi Xiu-Li, Lu Meng, Xiao Bin, Li Wei-Sheng. Pancreas segmentation based on dual-decoding U-Net. Journal of Software, 2022, 33(5): 1947−1958 [5] Rayed E, Islam S M S, Niha S I, Jim J R, Kabir M, Mridha M F. Deep learning for medical image segmentation: State-of-the-art advancements and challenges. Informatics in Medicine Unlocked, 2024, 47: Article No. 101504 doi: 10.1016/j.imu.2024.101504 [6] Li Z W, Liu F, Yang W J, Peng S H, Zhou J. A survey of convolutional neural networks: Analysis, applications, and prospects. IEEE Transactions on Neural Networks and Learning Systems, 2022, 33(12): 6999−7019 doi: 10.1109/TNNLS.2021.3084827 [7] Yao X J, Wang X Y, Wang S H, Zhang Y D. A comprehensive survey on convolutional neural network in medical image analysis. Multimedia Tools and Applications, 2022, 81(29): 41361−41405 doi: 10.1007/s11042-020-09634-7 [8] Sarvamangala D R, Kulkarni R V. Convolutional neural networks in medical image understanding: A survey. Evolutionary Intelligence, 2022, 15(1): 1−22 doi: 10.1007/s12065-020-00540-3 [9] Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Boston, USA: IEEE, 2015. 3431−3440 [10] Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. In: Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention. Munich, Germany: Springer, 2015. 234−241 [11] 殷晓航, 王永才, 李德英. 基于U-Net结构改进的医学影像分割技术综述. 软件学报, 2021, 32(2): 519−550Yin Xiao-Hang, Wang Yong-Cai, Li De-Ying. Suvery of medical image segmentation technology based on U-Net structure improvement. Journal of Software, 2021, 32(2): 519−550 [12] Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez A N, et al. Attention is all you need. In: Proceedings of the 31st International Conference on Neural Information Processing Systems. Long Beach, USA: Curran Associates Inc., 2017. 6000−6010 [13] Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X H, Unterthiner T, et al. An image is worth 16×16 words: Transformers for image recognition at scale. In: Proceedings of the 9th International Conference on Learning Representations. arXiv preprint arXiv: 2010.11929, 2020. [14] Conze P H, Andrade-Miranda G, Singh V K, Jaouen V, Visvikis D. Current and emerging trends in medical image segmentation with deep learning. IEEE Transactions on Radiation and Plasma Medical Sciences, 2023, 7(6): 545−569 doi: 10.1109/TRPMS.2023.3265863 [15] Yao W J, Bai J J, Liao W, Chen Y H, Liu M J, Xie Y. From CNN to transformer: A review of medical image segmentation models. Journal of Imaging Informatics in Medicine, 2024, 37(4): 1529−1547 doi: 10.1007/s10278-024-00981-7 [16] Han K, Wang Y H, Chen H T, Chen X H, Guo J Y, Liu Z H, et al. A survey on vision transformer. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2023, 45(1): 87−110 doi: 10.1109/TPAMI.2022.3152247 [17] Parvaiz A, Khalid M A, Zafar R, Ameer H, Ali M, Fraz M M. Vision transformers in medical computer vision——A contemplative retrospection. Engineering Applications of Artificial Intelligence, 2023, 122: Article No. 106126 doi: 10.1016/j.engappai.2023.106126 [18] Kirillov A, Mintun E, Ravi N, Mao H Z, Rolland C, Gustafson L, et al. Segment anything. arXiv preprint arXiv: 2304.02643, 2023. [19] Mazurowski M A, Dong H Y, Gu H X, Yang J C, Konz N, Zhang Y X. Segment anything model for medical image analysis: An experimental study. Medical Image Analysis, 2023, 89: Article No. 102918 doi: 10.1016/j.media.2023.102918 [20] He S, Bao R N, Li J P, Grant P E, Ou Y M. Accuracy of segment-anything model (SAM) in medical image segmentation tasks. arXiv preprint arXiv: 2304.09324v1, 2023. [21] Zhang K D, Liu D. Customized segment anything model for medical image segmentation. arXiv preprint arXiv: 2304.13785, 2023. [22] Xiao H G, Li L, Liu Q Y, Zhu X H, Zhang Q H. Transformers in medical image segmentation: A review. Biomedical Signal Processing and Control, 2023, 84: Article No. 104791 doi: 10.1016/j.bspc.2023.104791 [23] Chen J N, Lu Y Y, Yu Q H, Luo X D, Adeli E, Wang Y, et al. TransUNet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv: 2102.04306, 2021. [24] Wang W X, Chen C, Ding M, Yu H, Zha S, Li J Y. TransBTS: Multimodal brain tumor segmentation using transformer. In: Proceedings of the 24th International Conference on Medical Image Computing and Computer Assisted Intervention. Strasbourg, France: Springer, 2021. 109−119 [25] Wang H N, Cao P, Wang J Q, Zaiane O R. UCTransNet: Rethinking the skip connections in U-Net from a channel-wise perspective with transformer. In: Proceedings of the 36th AAAI Conference on Artificial Intelligence. California, USA: AAAI Press, 2022. 2441−2449 [26] Xie E Z, Wang W H, Yu Z D, Anandkumar A, Alvarez J M, Luo P. SegFormer: Simple and efficient design for semantic segmentation with transformers. In: Proceedings of the 35th International Conference on Neural Information Processing Systems. New Orleans, USA: Curran Associates Inc., 2021. Article No. 924 [27] Rahman M, Marculescu R. Medical image segmentation via cascaded attention decoding. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. Waikoloa, USA: IEEE, 2023. 6222−6231 [28] Cao H, Wang Y Y, Chen J, Jiang D S, Zhang X P, Tian Q, et al. Swin-Unet: Unet-like pure transformer for medical image segmentation. In: Proceedings of European Conference on Computer Vision——ECCV 2022 Workshops. Tel Aviv, Israel: Springer, 2023. 205−218 [29] Valanarasu J M J, Oza P, Hacihaliloglu I, Patel V M. Medical transformer: Gated axial-attention for medical image segmentation. In: Proceedings of the 24th International Conference on Medical Image Computing and Computer Assisted Intervention. Strasbourg, France: Springer, 2021. 36−46 [30] Huang X H, Deng Z F, Li D D, Yuan X G, Fu Y. MISSFormer: An effective transformer for 2D medical image segmentation. IEEE Transactions on Medical Imaging, 2023, 42(5): 1484−1494 doi: 10.1109/TMI.2022.3230943 [31] Li Z H, Zheng Y, Shan D D, Yang S Z, Li Q D, Wang B Z, et al. ScribFormer: Transformer makes CNN work better for scribble-based medical image segmentation. IEEE Transactions on Medical Imaging, 2024, 43(6): 2254−2265 doi: 10.1109/TMI.2024.3363190 [32] Yao M, Zhang Y Z, Liu G F, Pang D. SSNet: A novel transformer and CNN hybrid network for remote sensing semantic segmentation. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 2024, 17: 3023−3037 doi: 10.1109/JSTARS.2024.3349657 [33] Xu G A, Jia W J, Wu T, Chen L G, Gao G W. HAFormer: Unleashing the power of hierarchy-aware features for lightweight semantic segmentation. IEEE Transactions on Image Processing, 2024, 33: 4202−4214 doi: 10.1109/TIP.2024.3425048 [34] Panayides A S, Amini A, Filipovic N D, Sharma A, Tsaftaris S A, Young A, et al. AI in medical imaging informatics: Current challenges and future directions. IEEE Journal of Biomedical and Health Informatics, 2020, 24(7): 1837−1857 doi: 10.1109/JBHI.2020.2991043 [35] Liu Z, Lin Y T, Cao Y, Hu H, Wei Y X, Zhang Z, et al. Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. Montreal, Canada: IEEE, 2021. 10012−10022 [36] Dong X Y, Bao J M, Chen D D, Zhang W M, Yu N H, Yuan L, et al. CSWin transformer: A general vision transformer backbone with cross-shaped windows. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. New Orleans, USA: IEEE, 2022. 12124−12134 [37] Yang J W, Li C Y, Dai X Y, Gao J F. Focal modulation networks. In: Proceedings of the 36th International Conference on Neural Information Processing Systems. New Orleans, USA: Curran Associates Inc., 2022. Article No. 304 [38] Woo S, Debnath S, Hu R H, Chen X L, Liu Z, Kweon I S, et al. ConvNeXt V2: Co-designing and scaling ConvNets with masked autoencoders. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. Vancouver, Canada: IEEE, 2023. 16133−16142 [39] Luo X D, Liao W J, Xiao J H, Chen J N, Song T, Zhang X F, et al. WORD: A large scale dataset, benchmark and clinical applicable study for abdominal organ segmentation from CT image. Medical Image Analysis, 2022, 82: Article No. 102642 doi: 10.1016/j.media.2022.102642 [40] Synapse多器官分割数据集 [Online], available: https://www.synapse.org/#!Synapse:syn3193805/wiki/89480, 2023-05-13Synapse multi-organ segmentation dataset [Online], available: https://www.synapse.org/#!Synapse:syn3193805/wiki/89480, May 13, 2023 [41] Oktay O, Schlemper J, le Folgoc L, Lee M, Heinrich M, Misawa K, et al. Attention U-Net: Learning where to look for the pancreas. arXiv preprint arXiv: 1804.03999, 2018. [42] Yu Q, Qi L, Gao Y, Wang W Z, Shi Y H. Crosslink-Net: Double-branch encoder network via fusing vertical and horizontal convolutions for medical image segmentation. IEEE Transactions on Image Processing, 2022, 31: 5893−5908 doi: 10.1109/TIP.2022.3203223 [43] Chen S C, Ding C X, Liu M F, Cheng J, Tao D C. CPP-Net: Context-aware polygon proposal network for nucleus segmentation. IEEE Transactions on Image Processing, 2023, 32: 980−994 doi: 10.1109/TIP.2023.3237013 [44] He Z Q, Unberath M, Ke J, Shen Y Q. TransNuSeg: A lightweight multi-task transformer for nuclei segmentation. In: Proceedings of the 26th International Conference on Medical Image Computing and Computer Assisted Intervention. Vancouver, Canada: Springer, 2023. 206−215 [45] Li H P, Liu D R, Zeng Y, Liu S C, Gan T, Rao N N, et al. Single-image-based deep learning for segmentation of early esophageal cancer lesions. IEEE Transactions on Image Processing, 2024, 33: 2676−2688 doi: 10.1109/TIP.2024.3379902 [46] Hong Z F, Chen M Z, Hu W J, Yan S Y, Qu A P, Chen L N, et al. Dual encoder network with transformer-CNN for multi-organ segmentation. Medical and Biological Engineering and Computing, 2023, 61(3): 661−671 [47] Shao Y Q, Zhou K Y, Zhang L C. CSSNet: Cascaded spatial shift network for multi-organ segmentation. Computers in Biology and Medicine, 2024, 179: Article No. 107955 [48] Seidlitz S, Sellner J, Odenthal J, Özdemir B, Studier-Fischer A, Knödler S, et al. Robust deep learning-based semantic organ segmentation in hyperspectral images. Medical Image Analysis, 2022, 80: Article No. 102488 doi: 10.1016/j.media.2022.102488 [49] Gravetter F J, Wallnau L B, Forzano L A B, Witnauer J E. Essentials of Statistics for the Behavioral Sciences (Tenth edition). Australia: Cengage Learning, 2021. 326−333 [50] Zhou H Y, Gou J S, Zhang Y H, Han X G, Yu L Q, Wang L S, et al. nnFormer: Volumetric medical image segmentation via a 3D transformer. IEEE Transactions on Image Processing, 2023, 32: 4036−4045 doi: 10.1109/TIP.2023.3293771 [51] Liu Z, Mao H Z, Wu C Y, Feichtenhofer C, Darrell T, Xie S N. A ConvNet for the 2020s. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. New Orleans, USA: IEEE, 2022. 11976−11986 -




 下载:
下载: