-
摘要: 在医学图像中, 器官或病变区域的精准分割对疾病诊断等临床应用有着至关重要的作用, 然而分割模型的训练依赖于大量标注数据. 为减少对标注数据的需求, 本文主要研究针对医学图像分割的半监督学习任务. 现有半监督学习方法广泛采用平均教师模型, 其缺点在于, 基于指数移动平均(Exponential moving average, EMA)的参数更新方式使得老师模型累积学生模型的错误知识. 为避免上述问题, 提出一种双模型交互学习方法, 引入像素稳定性判断机制, 利用一个模型中预测结果更稳定的像素监督另一个模型的学习, 从而缓解了单个模型的错误经验的累积和传播. 提出的方法在心脏结构分割、肝脏肿瘤分割和脑肿瘤分割三个数据集中取得优于前沿半监督方法的结果. 在仅采用30%的标注比例时, 该方法在三个数据集上的戴斯相似指标(Dice similarity coefficient, DSC)分别达到89.13%, 94.15%, 87.02%.Abstract: Accurate segmentation of organs or lesions in medical images plays a significant role in clinical applications such as clinical diagnosis. However, learning segmentation models require a large number of annotated samples. This paper focuses on the semi-supervised medical image segmentation to relieve the dependence on labeled samples. A widely used semi-supervised learning method is temporally averaging a student model as the teacher model. However, it accumulates the incorrect knowledge of the student model as well. To address the above issue, we propose an interactive dual-model learning algorithm. Aiming to prevent the propagation and accumulation of error knowledge, we devise a specific mechanism for judging and measuring the instability of network predictions. Only pixels with relatively more stable predictions in one model are employed to supervise the other model. Extensive experiments on three datasets including cardiac structure segmentation, liver tumor segmentation, and brain tumor segmentation, demonstrate that the proposed method outperforms the state-of-the-art semi-supervised methods. When 30% of annotations are available, the Dice similarity coefficient (DSC) metric of our method reaches 89.13%, 94.15% and 87.02% respectively on the above three datasets.
-
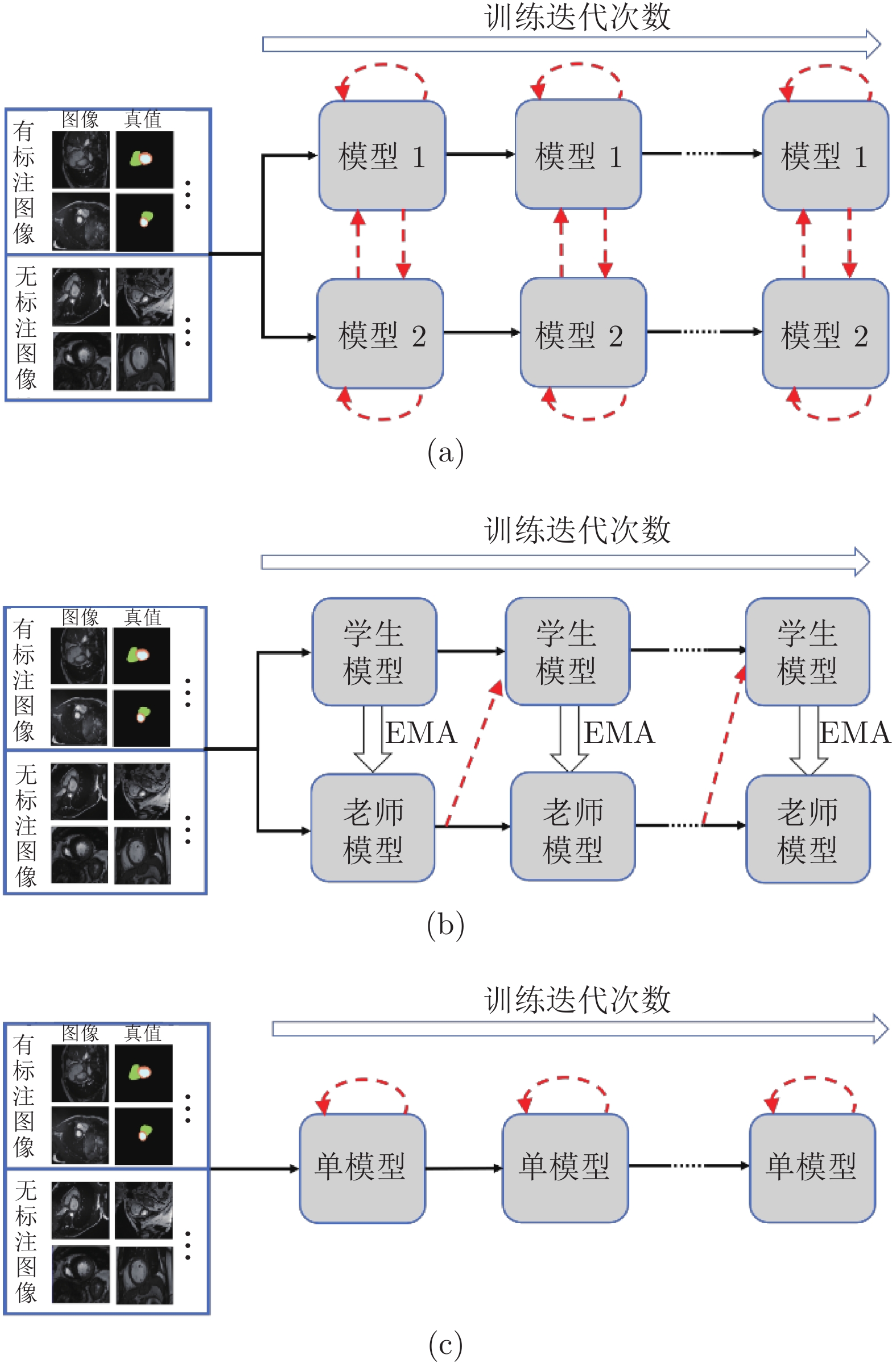
图 1 模型框架的对比图 ((a)基于双模型交互学习的半监督分割框架; (b)基于平均教师模型[22]的半监督分割框架; (c)基于一致性约束的单模型半监督分割框架. 实线箭头表示训练数据的传递和模型的更新, 虚线箭头表示无标注数据监督信息的来源)
Fig. 1 Comparison of the model framework ((a) Semi-supervised segmentation framework based on dual-model interactive learning; (b) Semi-supervised segmentation framework based on the mean teacher model[22]; (c) Semi-supervised segmentation framework based on single model. Solid arrows represent the propagation of training data and the update of models. Dashed arrows point out the origin of the supervisions on unlabeled images)
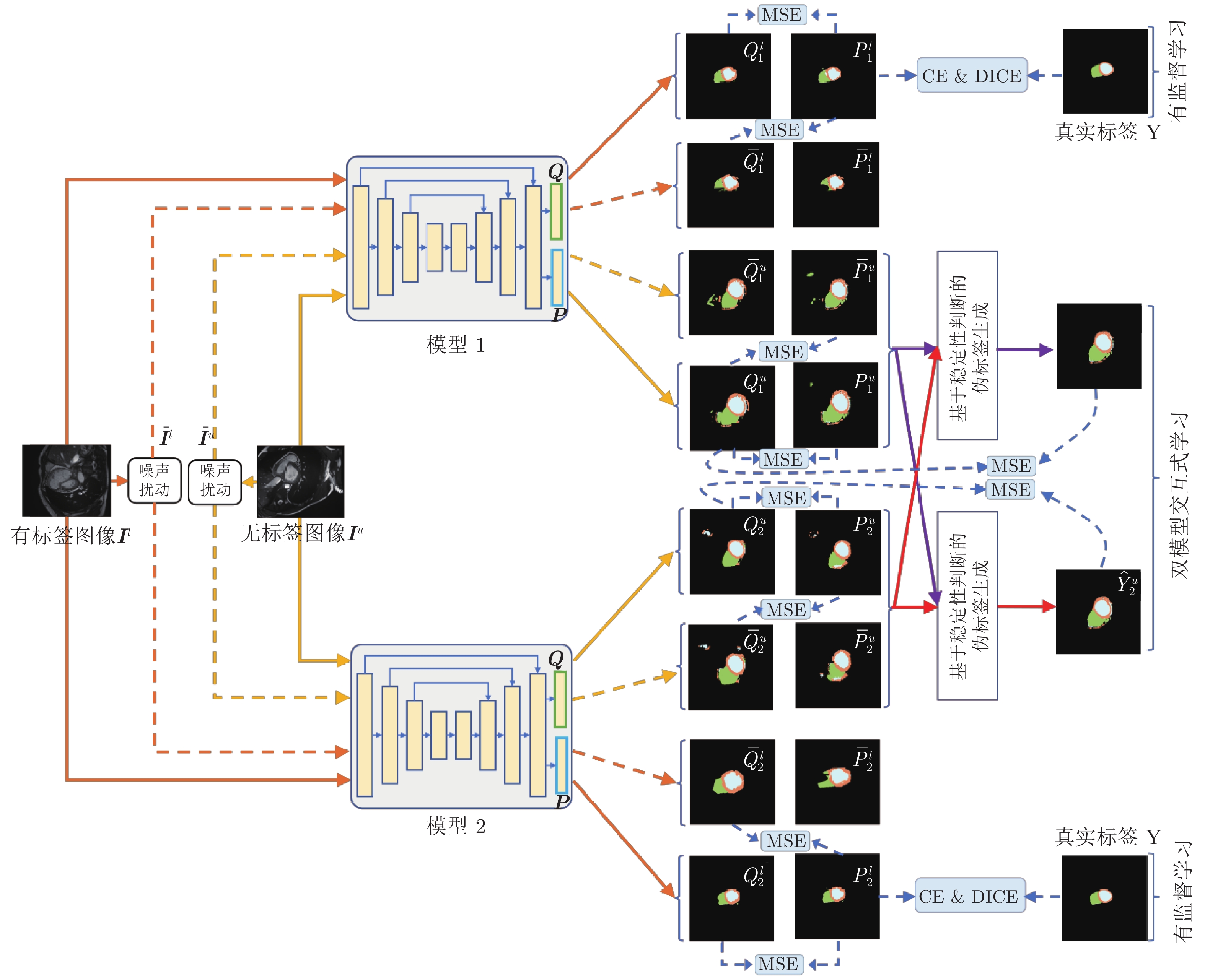
图 2 双模型交互学习框架图. MSE、CE 和 DICE 分别表示均方误差函数、交叉熵函数和戴斯函数. 单向实线箭头表示原始图像(
$ {{\boldsymbol{I}}}^{{{l}}} $ 和$ {{\boldsymbol{I}}}^{{{u}}} $ )在各模型中的前向计算过程, 单向虚线箭头表示噪声图像($ {{\bar{{\boldsymbol{I}}}}}^{{{l}}} $ 和$ {{\bar{{\boldsymbol{I}}}}}^{{{u}}} $ )在各模型中的前向计算过程Fig. 2 Framework of interactive learning of dual-models. MSE, CE and DICE represent mean square error function, cross entropy function and DICE function, respectively. The solid single-directional arrow represents the forward calculation process of the original image (
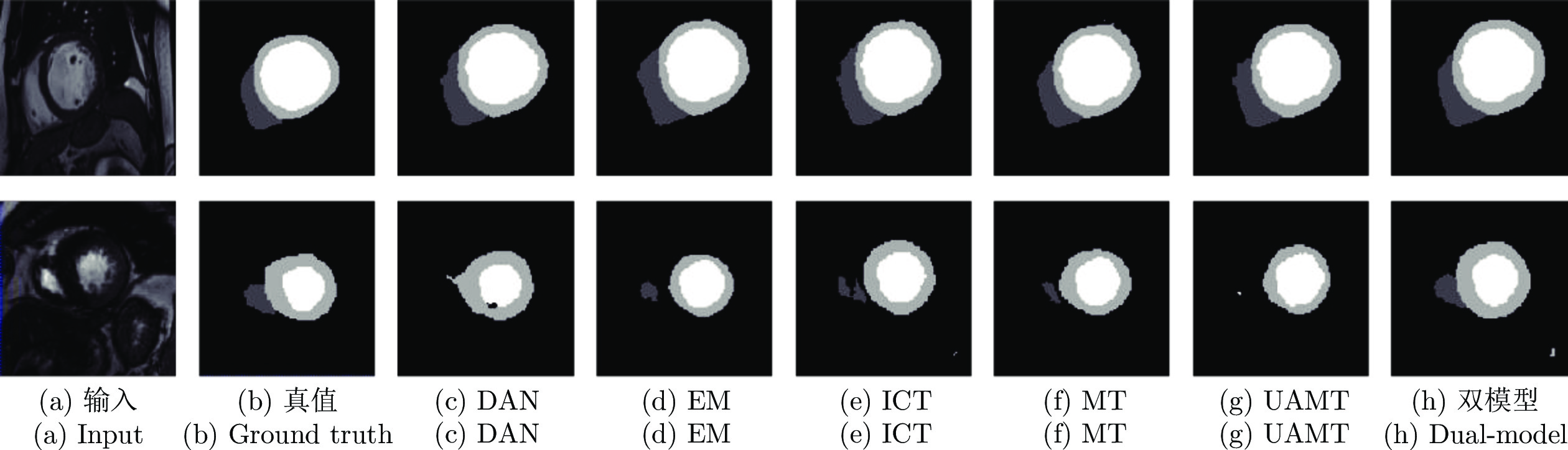
$ {{\boldsymbol{I}}}^{{{l}}} $ and$ {{\boldsymbol{I}}}^{{{u}}} $ ) in each model. The dashed single-directional arrow represents the forward calculation process of noise images (${{\bar{{{{\boldsymbol{I}}}}}}}^{{{l}}}$ and${{\bar{{\boldsymbol{I}}}}}^{{{u}}}$ ) in each model图 3 在 CSS 数据集中, 双模型与其他半监督方法分割结果图, 图中黑色区域代表背景, 深灰色区域代表左室腔,浅灰色区域代表左室心肌, 白色区域代表右室腔
Fig. 3 Segmentation results of our method and other semi-supervised methods on the CSS dataset. The black, dark gray, light gray, and white represents the background, left ventricle cavity (LV Cavity), left ventricular myocardium (LV Myo), and right ventricle cavity (RV Cavity), respectively
表 1 本文双模型方法与其他双模型方法的比较
Table 1 Compared with other dual-model methods
方法 任务 网络 损失函数 主要贡献 DML[29] 图像分类 残差网络
移动网络
宽残差网络
谷歌网络相对熵损失函数
交叉熵损失函数提出双模型, 两个小网络实现交互学习. 用 KL 散度评估两个模型网络预测结果之间的差异 FML[52] 图像分类 残差网络
宽残差网络相对熵损失函数
交叉熵损失函数
对抗损失函数提出双模型, 在 DML 基础上, 在两个网络模型输出预测结果之间引入对抗学习 D-N[53] 图像分类 计算机视觉组网络
残差网络交叉熵损失函数
学生与老师之间的知识提取损失函数提出双模型, 每个模型提取特征并通过辅助分类器做出预测.同时将两个分支提取的特征进行融合, 通过融合分类器得到整体分类结果 本文方法 半监督医学
图像分割U 形网络
密集 U 形网络
三维 U 形网络交叉熵损失函数
戴斯损失函数
均方误差函数提出双模型, 引入稳定伪标签判断机制, 用一个模型的稳定像素约束另一个模型的不稳定像素 表 2 采用U-Net和DenseU-Net网络结构时, 在不同标签比例的CSS数据集下与其他方法的对比结果
Table 2 Comparison with other methods on the CSS dataset when different training images are annotated. The baseline segmentation network is U-Net or DenseU-Net
基准模型 方法 5% 10% 20% 30% 50% DSC (%) HD95 ASD DSC (%) HD95 ASD DSC (%) HD95 ASD DSC (%) HD95 ASD DSC (%) HD95 ASD U-Net MT[28] 57.98 35.81 13.71 80.70 10.75 2.93 85.32 7.63 2.25 87.40 6.77 1.85 88.65 5.60 1.62 DAN[25] 53.82 35.72 14.61 79.35 9.64 2.69 84.67 7.56 2.28 86.31 6.70 2.05 88.40 3.69 1.10 TCSM[48] 50.82 23.26 9.02 79.71 14.27 3.59 85.51 6.90 1.95 87.21 7.10 1.77 89.03 5.19 1.46 EM[47] 59.95 12.68 3.77 82.28 8.32 2.56 84.72 7.60 2.36 87.75 5.98 1.89 89.12 9.92 2.51 UAMT[21] 55.08 25.24 8.23 80.04 8.04 2.21 84.85 6.35 1.99 87.52 6.59 2.01 89.30 4.76 1.30 ICT[30] 53.75 14.46 4.95 81.36 8.66 2.40 85.68 6.80 2.10 88.17 5.67 1.45 89.45 4.78 1.57 DS[51] 68.18 5.78 1.54 81.85 6.26 2.06 87.07 5.57 1.39 88.18 5.82 1.38 89.24 3.37 0.97 DML[29] 59.92 10.96 2.16 77.23 5.61 1.98 82.61 8.41 3.07 86.48 8.29 1.89 88.20 3.86 1.21 FML[52] 60.13 10.01 1.84 79.96 5.48 1.90 83.04 7.87 2.89 87.13 7.28 1.69 88.27 3.67 1.19 D-N[53] 57.61 15.26 5.36 75.06 10.49 3.01 82.13 8.71 3.41 86.41 8.05 1.76 88.08 4.08 1.27 双模型 76.94 5.38 1.47 87.53 3.20 1.11 88.67 5.28 1.35 89.13 5.64 1.47 90.11 2.51 0.86 DenseU-Net MT[28] 51.91 34.69 11.56 75.19 19.39 5.57 83.62 9.56 3.06 86.97 5.20 1.45 88.24 4.24 1.55 UAMT[21] 59.73 23.33 7.07 78.20 12.95 3.66 83.12 10.03 3.04 87.07 5.84 2.05 88.08 5.12 1.57 ICT[30] 71.10 13.25 3.89 83.41 14.06 3.51 85.68 7.83 2.45 87.74 4.39 1.41 88.63 4.80 1.39 双模型 81.34 3.69 1.14 87.45 3.93 1.47 87.98 5.15 1.18 88.20 3.46 0.98 89.60 3.03 0.95 表 3 采用U-Net和DenseU-Net网络结构, 在30%标签比例的LiTS数据集下与其他方法的对比结果
Table 3 Comparison with other methods on LiTS when 30% training images are annotated. The baseline segmentation network is U-Net or DenseU-Net
网络结构 方法 DSC (%) HD95 ASD U-Net MT[28] 86.98 0.88 0.17 DAN[25] 86.15 2.54 0.62 TCSM[48] 84.77 0.96 0.20 EM[47] 87.21 0.70 0.17 UAMT[21] 85.69 0.97 0.20 ICT[30] 88.42 0.99 0.21 DS[51] 86.90 1.23 0.61 DML[29] 84.92 1.26 0.92 FML[52] 85.14 0.97 0.25 D-N[53] 84.17 1.33 0.95 双模型 94.15 0.09 0.03 DenseU-Net MT[28] 93.69 0.17 0.04 UAMT[31] 93.91 0.18 0.05 ICT[30] 93.90 0.11 0.04 双模型 94.43 0.12 0.05 表 4 采用3D U-Net网络, 在30%标签比例的BraTS数据集下与其他方法的对比结果
Table 4 Comparison with other methods on the BraTS dataset when 30% training images are annotated. The baseline network is 3D U-Net
表 5 采用U-Net网络, 在标签比例为10%的CSS数据上验证不同变体对结果的影响
Table 5 Performance of different variants of our method on the CSS dataset when 10% training images are annotated. The baseline segmentation network is U-Net
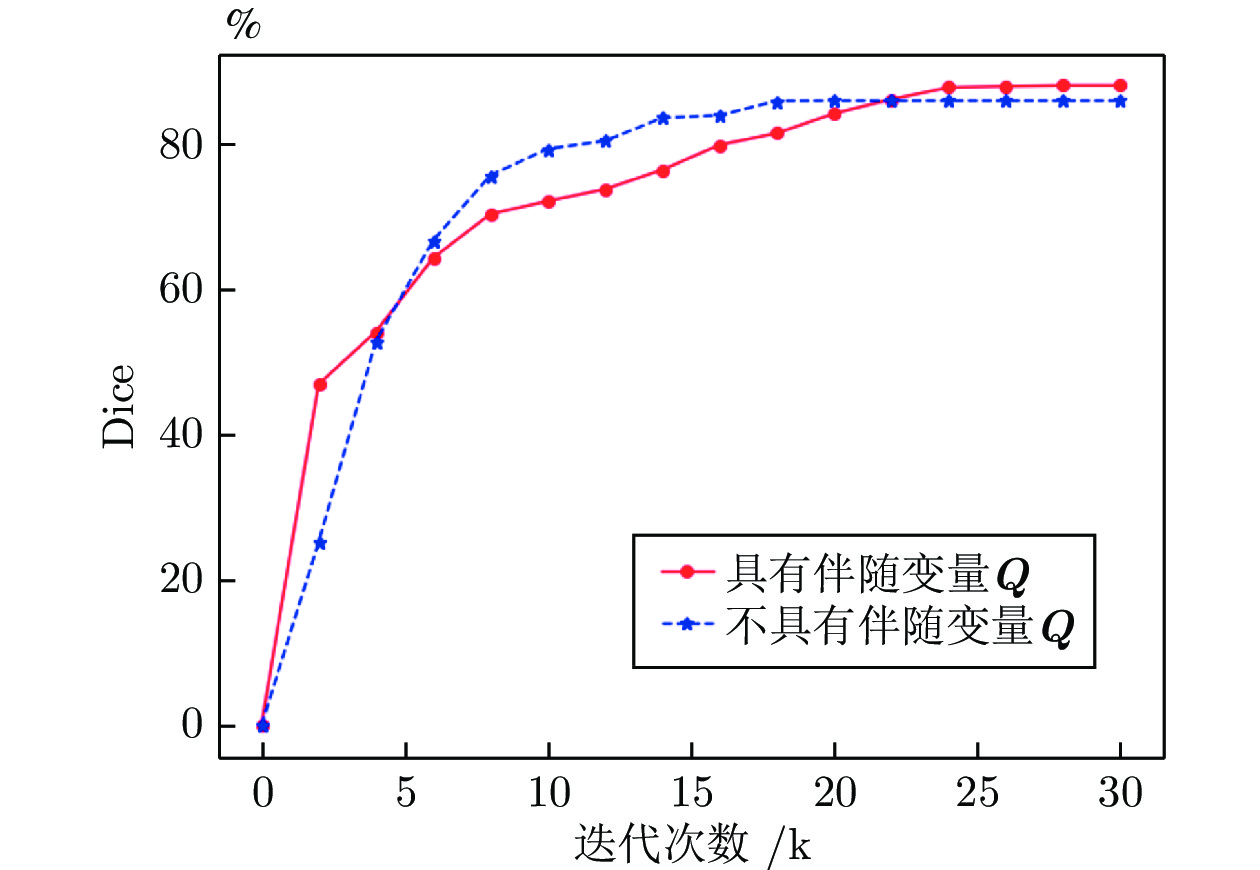
序号 有监督约束 无监督一致性 交互学习 稳定性选择策略 不使用伴随变量Q DSC (%) HD95 ASD 1 √ 76.41 10.46 3.12 2 √ √ 83.42 5.84 1.64 3 √ √ √ 85.52 5.47 1.57 4 √ √ √ 情况1) 86.62 4.92 1.44 5 √ √ √ 情况1)和情况2) √ 86.21 3.84 1.33 6 √ √ √ 情况1)和情况2) 87.53 3.20 1.11 表 6 采用U-Net网络, 在标签比例为10%的CSS数据上验证模型数量对结果的影响
Table 6 Performance of number of model on the CSS dataset when 10% training images are annotated. The baseline network is U-Net
学生数量 DSC (%) 2 87.53 4 87.32 6 87.46 表 7 采用U-Net网络, 在标签比例为10%的CSS数据上验证损失函数对结果的影响
Table 7 Performance of different loss function of our method on the CSS dataset when 10% training images are annotated. The baseline network is U-Net
损失函数 DSC (%) HD95 ASD ${L}_{{\rm{ce}}}$ 73.23 12.51 4.19 $ {L}_{{\rm{dice}}} $ 75.00 10.94 3.63 $ {L}_{{\rm{seg}}} $ 76.41 10.46 3.12 $ {L}_{{\rm{seg}}}+{L}_{{\rm{con}}\_P} $ 80.62 7.35 2.66 $ {L}_{{\rm{seg}}}+{L}_{{\rm{con}}\text{}} $ 83.42 5.84 1.64 ${L}_{ {\rm{seg} } }+{L}_{ {\rm{con} } }+{L}_{{\rm{sta}}}$ 87.53 3.20 1.61 表 8 采用U-Net网络, 在标签比例为10%的CSS数据集上验证损失函数对结果的影响
Table 8 Performance of network sharing of our method on the CSS dataset when 10% training images are annotated. The baseline network is U-Net
共享网络 DSC (%) HD95 ASD 单模型 83.04 6.02 4.13 编码器 85.05 4.41 2.65 双模型 87.53 3.20 1.61 -
[1] Krizhevsky A, Sutskever I, Hinton G E. ImageNet classification with deep convolutional neural networks. In: Proceedings of the 25th International Conference on Neural Information Processing Systems. Lake Tahoe, Nevada, USA: ACM, 2012. 1097−1105 [2] 罗建豪, 吴建鑫. 基于深度卷积特征的细粒度图像分类研究综述. 自动化学报, 2017, 43(8): 1306-1318Luo Jian-Hao, Wu Jian-Xin. A survey on fine-grained image categorization using deep convolutional features. Acta Automatica Sinica, 2017, 43(8): 1306-1318 [3] He K M, Zhang X Y, Ren S Q, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, USA: IEEE, 2016. 770−778 [4] Zhang D W, Zeng W Y, Yao J R, Han J W. Weakly supervised object detection using proposal- and semantic-level relationships. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2022, 44(6): 3349-3363 doi: 10.1109/TPAMI.2020.3046647 [5] Zhang D W, Han J W, Guo G Y, Zhao L. Learning object detectors with semi-annotated weak labels. IEEE Transactions on Circuits and Systems for Video Technology, 2019, 29(12): 3622-3635 doi: 10.1109/TCSVT.2018.2884173 [6] 刘小波, 刘鹏, 蔡之华, 乔禹霖, 王凌, 汪敏. 基于深度学习的光学遥感图像目标检测研究进展. 自动化学报, 2021, 47(9): 2078-2089Liu Xiao-Bo, Liu Peng, Cai Zhi-Hua, Qiao Yu-Lin, Wang Ling, Wang Min. Research progress of optical remote sensing image object detection based on deep learning. Acta Automatica Sinica, 2021, 47(9): 2078-2089 [7] Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Boston, USA: IEEE, 2015. 3431−3440 [8] Badrinarayanan V, Kendall A, Cipolla R. SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2017, 39(12): 2481-2495 doi: 10.1109/TPAMI.2016.2644615 [9] Li S L, Zhang C Y, He X M. Shape-aware semi-supervised 3D semantic segmentation for medical images. In: Proceedings of the 23rd International Conference on Medical Image Computing and Computer Assisted Intervention. Lima, Peru: Springer, 2020. 552−561 [10] Fang C W, Li G B, Pan C W, Li Y M, Yu Y Z. Globally guided progressive fusion network for 3D pancreas segmentation. In: Proceedings of the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention. Shenzhen, China: Springer, 2019. 210−218 [11] Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. In: Proceedings of the 18th International Conference on Medical Image Computing and Computer Assisted Intervention. Munich, Germany: Springer, 2015. 234−241 [12] Zhou Z W, Siddiquee M M R, Tajbakhsh N, Liang J M. UNet++: A nested U-Net architecture for medical image segmentation. In: Proceedings of the 4th International Workshop on Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Granada, Spain: Springer, 2018. 3−11 [13] 田娟秀, 刘国才, 谷珊珊, 鞠忠建, 刘劲光, 顾冬冬. 医学图像分析深度学习方法研究与挑战. 自动化学报, 2018, 44(3): 401-424Tian Juan-Xiu, Liu Guo-Cai, Gu Shan-Shan, Ju Zhong-Jian, Liu Jin-Guang, Gu Dong-Dong. Deep learning in medical image analysis and its challenges. Acta Automatica Sinica, 2018, 44(3): 401-424 [14] Zhu J W, Li Y X, Hu Y F, Ma K, Zhou S K, Zheng Y F. Rubik's Cube+: A self-supervised feature learning framework for 3D medical image analysis. Medical Image Analysis, 2020, 64: Article No. 101746 [15] Dai J F, He K M, Sun J. BoxSup: Exploiting bounding boxes to supervise convolutional networks for semantic segmentation. In: Proceedings of the IEEE International Conference on Computer Vision. Santiago, Chile: IEEE, 2015. 1635−1643 [16] Lin D, Dai J F, Jia J Y, He K M, Sun J. ScribbleSup: Scribble-supervised convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, USA: IEEE, 2016. 3159−3167 [17] Lee J, Kim E, Lee S, Lee J, Yoon S. FickleNet: Weakly and semi-supervised semantic image segmentation using stochastic inference. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. Long Beach, USA: IEEE, 2019. 5262−5271 [18] Chen C, Dou Q, Chen H, Heng P A. Semantic-aware generative adversarial nets for unsupervised domain adaptation in chest X-ray segmentation. In: Proceedings of the 9th International Workshop on Machine Learning in Medical Imaging. Granada, Spain: Springer, 2018. 143−151 [19] Ghafoorian M, Mehrtash A, Kapur T, Karssemeijer N, Marchiori E, Pesteie M, et al. Transfer learning for domain adaptation in MRI: Application in brain lesion segmentation. In: Proceedings of the 20th International Conference on Medical Image Computing and Computer Assisted Intervention. Quebec City, Canada: Springer, 2017. 516−524 [20] Li X M, Yu L Q, Chen H, Fu C W, Xing L, Heng P A. Semi-supervised skin lesion segmentation via transformation consistent self-ensembling model. In: Proceedings of the 29th British Machine Vision Conference. Newcastle, UK: BMVC, 2018. [21] Yu L Q, Wang S J, Li X M, Fu C W, Heng P A. Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation. In: Proceedings of the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention. Shenzhen, China: Springer, 2019. 605−613 [22] Nie D, Gao Y Z, Wang L, Shen D G. ASDNet: Attention based semi-supervised deep networks for medical image segmentation. In: Proceedings of the 21st International Conference on Medical Image Computing and Computer Assisted Intervention. Granada, Spain: Springer, 2018. 370−378 [23] Miyato T, Maeda S I, Koyama M, Ishii S. Virtual adversarial training: A regularization method for supervised and semi-supervised learning. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2019, 41(8): 1979-1993 doi: 10.1109/TPAMI.2018.2858821 [24] Laine S, Aila T. Temporal ensembling for semi-supervised learning. In: Proceedings of the International Conference on Learning Representations. Toulon, France: ICLR, 2017. [25] Zhang Y Z, Yang L, Chen J X, Fredericksen M, Hughes D P, Chen D Z. Deep adversarial networks for biomedical image segmentation utilizing unannotated images. In: Proceedings of the 20th International Conference on Medical Image Computing and Computer Assisted Intervention. Quebec City, Canada: Springer, 2017. 408−416 [26] Zheng H, Lin L F, Hu H J, Zhang Q W, Chen Q Q, Iwamoto Y, et al. Semi-supervised segmentation of liver using adversarial learning with deep atlas prior. In: Proceedings of the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention. Shenzhen, China: Springer, 2019. 148−156 [27] Ouali Y, Hudelot C, Tami M. Semi-supervised semantic segmentation with cross-consistency training. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. Seattle, USA: IEEE, 2020. 12671−12681 [28] Tarvainen A, Valpola H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. In: Proceedings of the 31st International Conference on Neural Information Processing Systems. Long Beach, USA: ACM, 2017. 1195−1204 [29] Zhang Y, Xiang T, Hospedales T M, Lu H C. Deep mutual learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. Salt Lake City, USA: IEEE, 2018. 4320−4328 [30] Verma V, Kawaguchi K, Lamb A, Kannala J, Solin A, Bengio Y, et al. Interpolation consistency training for semi-supervised learning. Neural Networks, 2022, 145: 90-106 doi: 10.1016/j.neunet.2021.10.008 [31] Luo X D, Chen J N, Song T, Wang G T. Semi-supervised medical image segmentation through dual-task consistency. In: Proceedings of the 35th AAAI Conference on Artificial Intelligence. Palo Alto, USA: AAAI, 2021. 8801−8809 [32] Cui W H, Liu Y L, Li Y X, Guo M H, Li Y M, Li X L, et al. Semi-supervised brain lesion segmentation with an adapted mean teacher model. In: Proceedings of the 26th International Conference on Information Processing in Medical Imaging. Hong Kong, China: Springer, 2019. 554−565 [33] Bernard O, Lalande A, Zotti C, Cervenansky F, Yang X, Heng P A, et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Transactions on Medical Imaging, 2018, 37(11): 2514-2525 doi: 10.1109/TMI.2018.2837502 [34] Bilic P, Christ P F, Vorontsov E, Chlebus G, Chen H, Dou Q, et al. The liver tumor segmentation benchmark (LiTS). arXiv: 1901.04056, 2019. [35] Menze B H, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Transactions on Medical Imaging, 2015, 34(10): 1993-2024 doi: 10.1109/TMI.2014.2377694 [36] You X G, Peng Q M, Yuan Y, Cheung Y M, Lei J J. Segmentation of retinal blood vessels using the radial projection and semi-supervised approach. Pattern Recognition, 2011, 44(10-11): 2314-2324 doi: 10.1016/j.patcog.2011.01.007 [37] Portela N M, Cavalcanti G D C, Ren T I. Semi-supervised clustering for MR brain image segmentation. Expert Systems With Applications, 2014, 41(4): 1492-1497 doi: 10.1016/j.eswa.2013.08.046 [38] Kohl S A A, Romera-Paredes B, Meyer C, De Fauw J, Ledsam J R, Maier-Hein K H, et al. A probabilistic U-Net for segmentation of ambiguous images. In: Proceedings of the 32nd International Conference on Neural Information Processing Systems. Montréal, Canada: ACM, 2018. 6965−6975 [39] Zhang Y, Zhou Z X, David P, Yue X Y, Xi Z R, Gong B Q, et al. PolarNet: An improved grid representation for online LiDAR point clouds semantic segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. Seattle, USA: IEEE, 2020. 9598−9607 [40] Isola P, Zhu J Y, Zhou T H, Efros A A. Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Honolulu, USA: IEEE, 2017. 5967−5976 [41] Lin T Y, Dollár P, Girshick R, He K M, Hariharan B, Belongie S. Feature pyramid networks for object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Honolulu, USA: IEEE, 2017. 936−944 [42] Li X M, Chen H, Qi X J, Dou Q, Fu C W, Heng P A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Transactions on Medical Imaging, 2018, 37(12): 2663-2674 doi: 10.1109/TMI.2018.2845918 [43] Milletari F, Navab N, Ahmadi S A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In: Proceedings of the 4th International Conference on 3D Vision. Stanford, USA: IEEE, 2016. 565−571 [44] Howard A G, Zhu M L, Chen B, Kalenichenko D, Wang W J, Weyand T, et al. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv: 1704.04861, 2017. [45] Ćićek Ö, Abdulkadir A, Lienkamp S S, Brox T, Ronneberger O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In: Proceedings of the 19th International Conference on Medical Image Computing and Computer-Assisted Intervention. Athens, Greece: Springer, 2016. 424−432 [46] Hang W L, Feng W, Liang S, Yu L Q, Wang Q, Choi K S, et al. Local and global structure-aware entropy regularized mean teacher model for 3D left atrium segmentation. In: Proceedings of the 23rd International Conference on Medical Image Computing and Computer Assisted Intervention. Lima, Peru: Springer, 2020. 562−571 [47] Vu T H, Jain H, Bucher M, Cord M, Pérez P. ADVENT: Adversarial entropy minimization for domain adaptation in semantic segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. Long Beach, USA: IEEE, 2019. 2512−2521 [48] Li X M, Yu L Q, Chen H, Fu C W, Xing L, Heng P A. Transformation-consistent self-ensembling model for semisupervised medical image segmentation. IEEE Transactions on Neural Networks and Learning Systems, 2021, 32(2): 523−534 [49] Yu X R, Han B, Yao J C, Niu G, Tsang I W, Sugiyama M. How does disagreement help generalization against label corruption? In: Proceedings of the 36th International Conference on Machine Learning. Long Beach, USA: PMLR, 2019. 7164−7173 [50] Han B, Yao Q M, Yu X R, Niu G, Xu M, Hu W H, et al. Co-teaching: Robust training of deep neural networks with extremely noisy labels. In: Proceedings of the 32nd International Conference on Neural Information Processing Systems. Montréal, Canada: ACM, 2018. 8536−8546 [51] Ke Z H, Wang D Y, Yan Q, Ren J, Lau R. Dual student: Breaking the limits of the teacher in semi-supervised learning. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. Seoul, South Korea: IEEE, 2019. 6727−6735 [52] Chung I, Park S, Kim J, Kwak N. Feature-map-level online adversarial knowledge distillation. In: Proceedings of the 37th International Conference on Machine Learning. Vienna, Austria: PMLR, 2020. 2006−2015 [53] Hou S H, Liu X, Wang Z L. DualNet: Learn complementary features for image recognition. In: Proceedings of the IEEE International Conference on Computer Vision. Venice, Italy: IEEE, 2017. 502−510 [54] Wang L C, Liu Y Y, Qin C, Sun G, Fu Y. Dual relation semi-supervised multi-label learning. In: Proceedings of the 34th AAAI Conference on Artificial Intelligence. New York, USA: AAAI, 2020. 6227−6234 [55] Xia Y C, Tan X, Tian F, Qin T, Yu N H, Liu T Y. Model-level dual learning. In: Proceedings of the 35th International Conference on Machine Learning. Stockholm, Sweden: PMLR, 2018. 5383−5392 -




 下载:
下载: